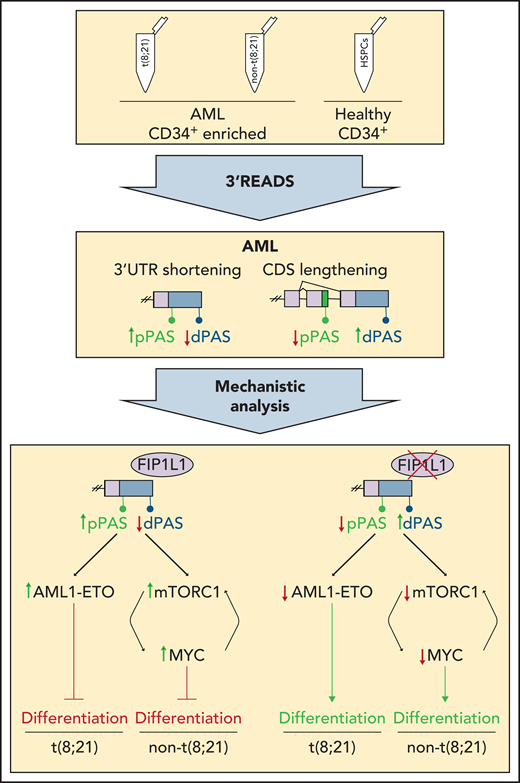
In this issue of Blood, Davis et al show that the posttranscriptional alternative polyadenylation (APA) process contributes to acute myeloid leukemia (AML) pathogenesis and that targeting the APA regulator FIP1L1 in AML cell lines promotes differentiation.1 The authors quantify changes in APA patterns by performing 3′ untranslated region (UTR) extraction and deep sequencing (3′ READS) (see figure) on samples from patients with primary AML and on hematopoietic stem and progenitor cells (HSPCs) from healthy donors. Mechanistic analysis suggests that targeting APA promotes differentiation of AML cells.
3′ READS was performed on human AML samples with and without t(8;21) that were enriched for CD34+ cells and on healthy CD34+ HSPCs to define global APA profiles. In AML samples, shortening of 3′ UTRs and lengthening of CDSs was detected. Mechanistic analysis revealed FIP1L1 as a major regulator of APA in AML that mediates global 3′ UTR shortening. In AML cells with t(8;21), this leads to increased levels of AML1-ETO, whereas in AML cells without t(8;21), mTORC1 and MYC levels are increased. This mechanism contributes to the characteristic block of AML cell differentiation. Knockdown of FIP1L1 reversed the characteristic APA patterns and induced differentiation of AML cells. dPAS, distal poly(A) site; pPAS, proximal poly(A) site.
3′ READS was performed on human AML samples with and without t(8;21) that were enriched for CD34+ cells and on healthy CD34+ HSPCs to define global APA profiles. In AML samples, shortening of 3′ UTRs and lengthening of CDSs was detected. Mechanistic analysis revealed FIP1L1 as a major regulator of APA in AML that mediates global 3′ UTR shortening. In AML cells with t(8;21), this leads to increased levels of AML1-ETO, whereas in AML cells without t(8;21), mTORC1 and MYC levels are increased. This mechanism contributes to the characteristic block of AML cell differentiation. Knockdown of FIP1L1 reversed the characteristic APA patterns and induced differentiation of AML cells. dPAS, distal poly(A) site; pPAS, proximal poly(A) site.
More than 70% of all genes harbor more than 1 polyadenylation site, and differential usage of these sites is termed APA.2 Depending on the exact localizations of the different polyadenylation sites, this process can affect the 3′ UTR as well as the coding DNA sequence (CDS). APA can influence RNA stability, RNA output, protein localization, and protein isoform expression. Occurring within the 3′ UTR, APA determines the length of the UTR, which directly affects the presence or absence of binding motifs and regulatory sequences. In contrast, differential usage of polyadenylation sites within exons, introns, and alternative 3′ UTRs leads to changes in the CDS of the transcript.
Large-scale sequencing of transcriptomes in different cancer entities and the application of novel 3′ sequencing methods have revealed that changes in APA are a common feature of malignant transformation.3 Although multiple studies have dealt with the prevalence of APA in solid tumors, few reports have focused on hematologic diseases and leukemia. Recently, APA was studied in healthy murine HSPCs and samples from patients with primary acute lymphoblastic leukemia, chronic lymphocytic leukemia, and AML.4-7 These studies suggest that APA plays a key role in the development of hematologic diseases. To date, however, mechanistic insights into how APA is involved in leukemia pathogenesis have remained elusive, which has also limited the potential for therapeutic exploitation. The study by Davis et al serves as a resource for APA patterns in samples from patients with AML and from HSPCs from healthy individuals, provides a comprehensive mechanistic analysis of the signaling pathways deregulated by APA in AML, and introduces APA as a putative target for AML therapy.
Davis et al performed 3′ READS on primary CD34+ AML blasts from patients with or without a t(8;21) translocation, which leads to the generation of an AML1-ETO fusion oncoprotein, using healthy CD34+ HSPCs as controls. Comparison of the data sets revealed global shortening of 3′ UTRs and lengthening of CDSs in samples from patients with AML. The genes affected by APA are involved in processes such as cell cycle, differentiation, and oncogenic signaling, and they include targets known for their role in blocking AML cell differentiation. Thus, several specific genes are deregulated by APA in AML and thereby potentially contribute to AML pathogenesis. Davis et al identified FIP1L1 as a global regulator of APA in AML. FIP1L1 is part of the polyadenylation machinery, and it promotes the use of proximal polyadenylation sites to maintain globally short 3′ UTRs.8 By using t(8;21) Kasumi cells as a model system, the authors demonstrated that knockdown of FIP1L1 reverses the AML APA phenotype, leading to global lengthening of 3′ UTRs and shortening of CDSs and accompanied by leukemic cell differentiation. This seminal result reveals that targeting of APA regulators can induce differentiation of AML cells. Mechanistically, FIP1L1 knockdown induces 3′ UTR lengthening of the AML1-ETO fusion transcript, which results in lower protein levels. Knockdown of FIP1L1 in AML cell lines without t(8;21) leads to attenuation of mTORC1 signaling and reduced levels of MYC protein, ultimately promoting differentiation and pointing to a broader role of FIP1L1 in regulating APA in AML.
The results by Davis et al are pivotal and provide a solid resource data set that can be exploited to identify additional targets and modulators of APA in AML. Of particular interest are the potential clinical applications. All-trans retinoic acid and arsenic trioxide have successfully been used in patients with acute promyelocytic leukemia, and isocitrate dehydrogenase (IDH) inhibitors have been used in IDH-mutant AML as pro-differentiation therapies. In the Davis et al study, targeting of APA regulators such as FIP1L1 is presented as an additional way to induce differentiation of AML cells. For clinical applications, the use of FIP1L1 inhibitors is conceivable. However, additional studies must be conducted to analyze potential adverse effects on healthy HSPCs. In the future, direct targeting of global APA patterns could represent an alternative to targeting APA regulators, and it might be beneficial in treating leukemia and other cancer entities. Different small molecules and morpholino oligomers have recently been shown to transiently regulate APA profiles, which opens new avenues for therapeutic uses.9,10
Overall, the study by Davis et al shows how APA deregulation can make AML cells more aggressive and immature by blocking their differentiation. Additional studies on APA in primary samples from other AML subtypes and leukemias may contribute to a comprehensive understanding of the functional role of APA in these cancers. It will be important to analyze bulk samples, to compare APA between leukemic stem cell and non–stem cell populations, and even perform single-cell resolution analysis. Because previous studies have shown both quantitative and qualitative changes in APA within differentiation hierarchies, these processes might also be differentially regulated between leukemic stem cell and blast fractions and could represent a druggable vulnerability that merits further investigation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal