In this issue of Blood, Yasuda et al identify 2 putative subgroups of acute lymphoblastic leukemia (ALL) in adolescents and adults driven by high CDX2 expression and IDH1/2 mutations that are associated with poor prognosis.1
B-precursor ALL (B-ALL) is a genetically heterogeneous disease characterized by distinct founding alterations, including aneuploidy, chromosomal rearrangements and sequence mutations that are important for risk stratification.2 Although ALL is less common in adolescents and young adults (AYAs) and adults than children, survival rates are markedly inferior (historically 30%-40% for adults compared with >85% for children), and the long-term prognosis for adults is poor.2 In recent years, significant advancements have been made in treating both AYAs and adults because in part to the increased use of pediatric-inspired chemotherapy regimens3 and the introduction of novel immune therapies, including monoclonal antibodies (eg, inotuzumab ozogamicin, blinatumomab) and chimeric antigen receptor T cells, which have shown remarkable activity in the relapsed/refractory setting and are now being tested in frontline trials.4 Improvements in our understanding of the biological differences in ALL across the age spectrum have also helped to explain this discrepant prognosis. In the era of cytogenetic diagnostic assays, it was observed that the prevalence of genetic alterations associated with a favorable outcome decreased in adults (eg, hyperdiploid ALL), whereas those associated with a poor prognosis simultaneously increased (eg, the Philadelphia chromosome [Ph]).2 More recent comprehensive next-generation sequencing has revealed multiple cytogenetically cryptic alterations, most commonly gene rearrangements, that constitute distinct ALL subgroups driven by distinct gene expression profiles.5 These studies have further refined the genomic landscape of AYA and adult ALL and have identified new therapeutic targets, with the identification of subtypes associated with poor outcome, such as Ph-like ALL, but have also revealed subsets of adult ALL patients with a relatively good prognosis, such as those harboring rearrangement of DUX4.6 They have also revealed new founding roles for genes previously considered support players in the development of ALL, including subgroups driven by PAX5 (PAX5 P80R and PAX5alt).5 Despite these new insights, a significant proportion of ALL remains genetically unclassified.
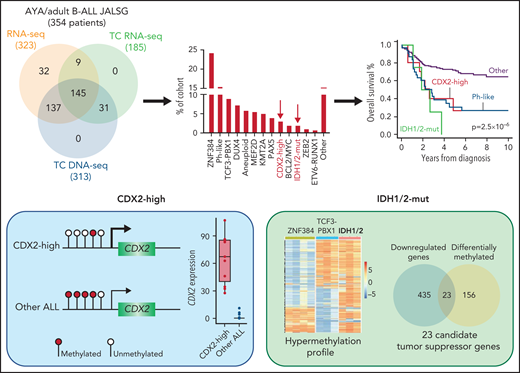
This study by Yasuda et al extends our knowledge of the genomic repertoire in ALL. They performed integrated whole transcriptomic and targeted DNA sequencing on a cohort of 354 Ph-negative B-ALL cases aged 15 to 64 years enrolled on consecutive Japan Adult Leukemia Study Group trials and classified them into 22 specific molecular subtypes (see figure). Two small but distinct clusters of patients were identified, characterized by high CDX2 expression (3.4%, “CDX2-high”) and IDH1/2 mutations (1.9%, “IDH1/2-mut”). The frequency of both groups was significantly lower in children: 0.3% and 0%, respectively. Of clinical importance, both CDX2-high and IDH1/2-mut subgroups were independently associated with poor survival, similar to Ph-like ALL.
An integrated analysis was performed on 354 adolescent and young adult (AYA) and adult patients with B-ALL enrolled in Japan Adult Leukemia Study Group (JALSG) trials. Genomic profiling included whole transcriptome sequencing (RNA-seq), targeted capture (TC) RNA-seq, and TC DNA sequencing. The authors identified 2 novel and distinct subgroups of B-ALL characterized by high CDX2 expression (3.4%, CDX2-high) and IDH1/2 mutations (1.9%, IDH1/2-mut) that were both associated with poor outcome. The promoter of CDX2 was hypomethylated in the CDX2-high group compared with other ALL, leading to outlier high expression. The IDH1/2-mut group was associated with a hypermethylation profile that converged on 23 candidate tumor suppressor genes that may play a role in IDH1/2-induced leukemogenesis.
An integrated analysis was performed on 354 adolescent and young adult (AYA) and adult patients with B-ALL enrolled in Japan Adult Leukemia Study Group (JALSG) trials. Genomic profiling included whole transcriptome sequencing (RNA-seq), targeted capture (TC) RNA-seq, and TC DNA sequencing. The authors identified 2 novel and distinct subgroups of B-ALL characterized by high CDX2 expression (3.4%, CDX2-high) and IDH1/2 mutations (1.9%, IDH1/2-mut) that were both associated with poor outcome. The promoter of CDX2 was hypomethylated in the CDX2-high group compared with other ALL, leading to outlier high expression. The IDH1/2-mut group was associated with a hypermethylation profile that converged on 23 candidate tumor suppressor genes that may play a role in IDH1/2-induced leukemogenesis.
The authors demonstrate outlier allele specific expression of CDX2 compared with all other ALL and normal lymphocytes that was associated with hypomethylation at the CDX2 promoter and recurrent gain of chromosome 1q. Expression of CDX2 at the protein level was confirmed in matched diagnosis and relapse samples, suggesting its importance in both the development and maintenance of leukemia. CDX2 is a caudal-related homeobox transcription factor involved in early embryogenesis and hematopoietic development, although its expression in adult tissues is restricted to the intestine.7 Although previous studies have demonstrated moderate levels of CDX2 expression in acute myeloid and lymphoid leukemia,8 this is the first study to demonstrate by whole transcriptome sequencing that outlier high expression constitutes a distinct ALL subgroup. Interestingly, in contrast to its role in normal hematopoietic development, CDX2-induced leukemogenesis is not driven by HOX genes in ALL but may involve different mechanisms including the insulin-like growth factor-1 receptor pathway, which also provides a potential therapeutic target.
The IDH1/2-mut subgroup was characterized by clonal IDH1 R132C and IDH2 R140Q mutations in diagnosis samples that persisted through to relapse in cases with available material. IDH1/2 genes are known initiating events in AML and brain tumors, but their role in ALL is less clear. The distinct gene expression signature, including upregulation of genes associated with mitochondrial function, and a paucity of additional genomic alterations in this new subgroup suggest that IDH1/2 may also be founding events in ALL. In AML, mutations in IDH1/2 are associated with increased methylation.9 Likewise, the IDH1/2-mut ALL group harbored a hypermethylation profile compared with other ALL, suggesting a new mechanism for lymphoid leukemogenesis, whereby aberrant methylation may lead to silencing of critical genes important for B-cell development. Accordingly, by combining DNA methylation and gene expression data, the authors identified 23 candidate tumor suppressor genes whose downregulation may play a role in IDH1/2-mut ALL. Functional experiments are required to validate these findings.
This paper lays the foundation for future studies focused on understanding the importance of CDX2 and IDH1/2 in the development of ALL, and significant work is required to demonstrate their role as initiating or founding events. For CDX2-high, the genomic mechanism responsible for outlier CDX2 expression and additional genomic alterations that are required for leukemogenesis are still unknown. Comprehensive whole genome sequencing or long-read sequencing platforms will be helpful in illuminating these underlying mechanisms. Understanding the biology of these subgroups and functional contribution of CDX2 and IDH1/2 to leukemogenesis is also a critical question that remains to be explored. Furthermore, ∼15% of AYA and adult ALL cases in this study remained uncharacterized; we must continue to push the boundaries of genomic technologies and algorithms to uncover all subtype-defining alterations in ALL.
Conflict-of-interest disclosure: The author declares no competing financial interests.

Comments
IDH1/2 mutations are associated with poor outcomes in adult Ph (-)- B-cell acute lymphoblastic leukemia
Kaiqi Liu b, d#, Xiaoyuan Gong b,c,Qiuyun Fang b,c, Chunlin Zhou b,d,Hui Wei a,b, c,d, Bingcheng Liu b, d, Yingchang Mi a, b, c,d,Jianxiang Wang a, b, c,d, Ying Wang a, b , c,d *
a. State Key Laboratory of Experimental Hematology,
b. National Clinical Research Center for Blood Disease
c. Haihe Laboratory of Cell Ecosystem,
d. Institute of Hematology and Blood Disease Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Tianjin, 300020, China.
Correspondence: Dr. Ying Wang. Email: wangying1@ihcams.ac.cn
To the Editor
We read the article by Yasuda T et al [1] published in 24 March 2022 on clinical impact of IDH1/2 mutations in acute lymphoblastic leukemia (ALL) with great interest. These data undoubtedly provide a new factor to predict poor prognosis in Ph- B-ALL and a new ALL subtype. As the IDH1/2 mutations often reported in AML, rare in ALL [2].
To complement their findings, we report outcomes of Ph- B-ALL with IDH1/2 mutation patients in our department. From November 2014 to December 2021, gene mutations of 283 untreated Ph- B-ALL patients were tested by NGS and 14 patients were found with IDH1/2 mutation (IDH1/2-mut, 4.95%). The mutation positions include IHD1 R132C (n=4), IDH1 R132S (n=2), IDH1 Y208C (n=3), IDH2 R140Q (n=3), IDH2 A347T (n=1), IDH2 M397V (n=1). Outside of IDH1 R132C and IDH2 R140Q mutation, the other 4 mutation sites in our patients were not reported in Yasuda T’s article. ALL 14 cases with IDH1/2-mut ALL patient, male/female was 1:1, median age 47 (16-69) years old, median WBC count was 2.7 (1.03-245.0)x109/L. 13 patients had chromosome karyotype results, 11 with normal karyotype, one with +Y, one with der(8;17)(q10;q10),der(9)?t(8;9)(q11.2;p10). 7 patients were diagnosed as common-B ALL and 7 cases pre-B ALL.
All the IDH1/2-mut patients was induced with VDC(L)P regimen according to the age of the patient. Complete response (CR) rates was 100%. All the patients were followed up until March 30, 2022. The overall survival (OS) between the IDH1/2-mut and IDH1/2-wild type (IDH1/2-wt) patients was compared (Figure1). The OS of IDH1/2-mut patients was inferior to that of IDH1/2-wt. 2 years OS was 32.5% (IDH1/2-mut) vs 63.6% (IDH1/2-wt). But there was no statistical difference between two groups (p=0.358), may due to the number of the patients with IDH1/2-mt is small.
In summary, we present a cohort of IDH1/2-mut Ph- B-ALL. IDH1/2 mutations didn’t affect the CR rate. But, the OS of IDH1/2-mut B-ALL may be inferior to that of IDH1/2-wt B-ALL consistent with Yasuda T reported. In recent years, clinical trials show that ivosidenib, enasidenib and venetoclax improved therapeutic effect of AML with IDH1/2 mutations. In the future, we may add these drugs to the treatment of ALL with IDH1/2 mutations.
Ethics approval and consent to participate:
All studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the Helsinki Declaration 2013. Written informed consent was obtained from the patients or patients’ parents/legal guardians.
Authorship Statement
Kaiqi Liu and Xiaoyuan Gong collected the data and wrote the paper. Qiuyun Fang , Chunlin Zhou ,Hui Wei,Bingcheng Liu, Yingchang Mi were involved in patient management and clinical data collection. Jianxiang Wang and Ying Wang revised the manuscript and contributed the valuable advice.
Conflicts of Interest
All the authors have reviewed and approved the manuscript and do not have any disclosures/conflicts of interest.
This study was supported by National Key Research and Development Program of China (2019YFC0840605), and Tianjin Municipal Science and Technology Commission Grant (20JCZDJC00120).
References:
1 Yasuda T, Sanada M, Kawazu M, et al. Two novel high-risk adult B-cell acute lymphoblastic leukemia subtypes with high expression of CDX2 and IDH1/2 mutations. Blood. 2022,139(12):1850-1862.
2 DiNardo CD, Ravandi F, Agresta S, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am. J. Hematol. 2015, 90: 732–736.
3 DiNardo CD, Stein AS, Stein EM, et al. Mutant isocitrate dehydrogenase 1 inhibitor ivosidenib in combination with azacitidine for newly diagnosed acute myeloid leukemia. J Clin Oncol. 2021, 39(1): 57-65.
4 Pollyea DA, Tallman MS, Botton SD, et al. Enasidenib, an inhibitor of mutant IDH2 proteins, induces durable remissions in older patients with newly diagnosed acute myeloid leukemia. Leukemia. 2019, 33: 2575–84.
5 DiNardo CD, Jonas BA, Pullarkat V, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020, 383(7): 617-629.