In this issue of Blood, Boldrin et al1 identify a novel biomarker of poor outcome in acute lymphoblastic leukemia (ALL), and delineate the biological link between miR-497/195 repression and more aggressive disease.
ALL is the exemplar cancer in which biological subclassification critically informs risk stratification and treatment selection. With evolution from G-band karyotyping and fluorescence in situ hybridization–based detection of structural abnormalities to molecular approaches, including whole-genome and RNA sequencing, 23 discrete subtypes of B-cell precursor ALL (BCP-ALL) have now been characterized by specific driver events and gene-expression profiles.2 More than an exercise in taxonomy, gene-expression profiling was pivotal in identifying tyrosine kinase–activating events that mimic BCR-ABL1, defining the Philadelphia chromosome–like subtype of BCP-ALL.3 Moreover, it is becoming evident that detailed molecular classification of ALL contributes to improved treatment outcomes.4
Boldrin et al now layer the expression of miR-497/195 onto the molecular-subtyping scaffold. They link the expression of these microRNAs (miRNAs) to clinical outcomes, showing that lower expression is independently associated with diminished event-free and relapse-free survival.
Identifying novel recurring aberrations provides insights into disease biology, more precise risk stratification, and the potential to design more efficacious therapy. Sequencing of large ALL cohorts and deconvoluting these complex data sets using probability-based algorithms has allowed recognition of increasingly rare driver mutations.2 Here, a candidate approach was instead used to identify the putative determinants of an informative phenotype; in this case, differential expression of miRNAs associated with time to engraftment in a cohort of leukemia xenografts that demonstrated phenotypic heterogeneity. Leukemias with known genetic lesions were deliberately excluded from the initial analysis to reduce possible confounding factors. Analysis of a wider cohort, including ALL with recognized mutations, then provided the opportunity to delineate the potential role of miR-497/195 expression as a risk modifier associated with or within defined disease subtypes. Identifying such biomarkers is of clinical interest, particularly if they help explain the heterogeneity in outcomes still observed within the currently defined risk groups.
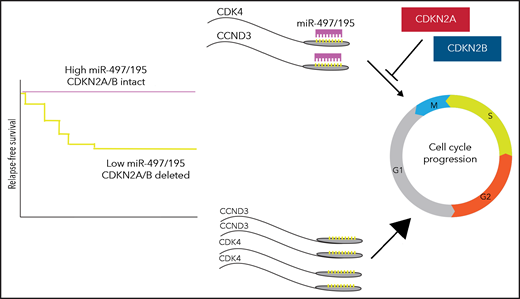
The authors have gone beyond description of a molecular association to map out a biologically plausible framework. They show that miR-497/195 normally represses cyclin-dependent kinase 4 (CDK4) and cyclin D3 (CCND3). Elevated CDK4 and CCND3 expression as a consequence of low miR-497/195 drives proliferation, an effect that is exaggerated when CDKN2A/CDKN2B, negative regulators of CDK4, are also deleted. The cooperative biology of concurrent miR-497/195 and CDKN2A/B mutations raises further interesting considerations. Highly deleterious mutations affecting pathways at different levels are often mutually exclusive, for example, TP53 and ARF, or KRAS and EGFR, as their co-occurrence confers no additional benefit to the cancer cell. However, as the mutational landscape of cancer is characterized with ever-increasing resolution, recurrent mutations with less pronounced or consistent biological and prognostic impacts are emerging. In ALL, the prognostic significance of CRLF2 mutations is highly context-dependent with adverse implications in patients with other high-risk features,5 particularly with concurrent JAK2 mutations, though not in standard-risk patients or on a background of trisomy 21.6 Similarly, although CDKN2A/B mutations are common and not generally associated with outcome, they modulate risk associated with IKZF1 mutations, constituting a defining lesion of the poor-prognosis IKZF1plus subgroup.7 This work may have identified another scenario in which CDKN2A/B mutations play an important cooperative role, with the synthetic mutational effect amplifying an adverse phenotype, notably here with both mutations acting in the same pathway (see figure).
Increased CDK4 and CCND3 expression, resulting from low miR-497/195, cooperate with loss of CDKN2A/B to drive greater cell proliferation.
Increased CDK4 and CCND3 expression, resulting from low miR-497/195, cooperate with loss of CDKN2A/B to drive greater cell proliferation.
In the face of increasing complexity and volume of data, what is the best approach to integrating combined mutational information into clinical practice? Although ALL risk stratification schema for major cooperative trial groups like the Children’s Oncology Group remain defined by individual lesions, there is an emerging case for considering the context of individually less-deleterious mutations and the prognostic significance of biological synergy. Defining disease subtypes by gene-expression profiles using RNA sequencing represents an already feasible means to capture this integrated impact.
The biology of leukemia is multifaceted, and aberrations that initiate leukemogenesis or drive clonal evolution may overlap with, or be distinct from, those important for disease persistence or treatment response. How the experimental end point that underpinned this work, time to engraftment in immunodeficient mice, relates to biological processes in ALL is worth considering. It may, for example, reflect the kinetics of tumor growth and greater self-sufficiency in growth signaling. Of note, however, miR-497/195 expression did not correlate with end-of-induction minimal residual disease (the most robust clinical measure of responsiveness to therapy), nor was there enrichment for low expression in comparisons between primary and relapse samples, suggesting that low miR-497/195 may not exert an adverse impact on outcome by promoting chemoresistance. Could greater proliferation potentially impact outcome purely through effects on relative disease burden and kinetics, necessitating deeper remission to achieve cure? Or does this hint at a more fundamental biological driver of poor outcome? For example, it has been demonstrated that increased cell proliferation independently drives tumorigenesis.8 Might this also promote greater tumor heterogeneity and potential for clonal evolution? Longitudinal single-cell sequencing approaches might provide further insights.
So, is miR-497/195 ready to be included in the risk stratification of BCP-ALL? The many orthogonal lines of evidence presented in the article, bringing together clinical observations, in vivo models, and functional genomics, make for a compelling story. Certainly, genomic techniques such as RNA sequencing are able to characterize miRNA expression while identifying driver fusion oncogenes, individual gene abundance, and gene-expression profiles. Moreover, the vast expansion of RNA-sequencing endeavors in BCP-ALL provides a wealth of reference data and retrospective analyses for further validation of this biomarker. Most importantly, the inclusion of RNA sequencing in current multicenter BCP-ALL trials provides the opportunity to prospectively test associations between miR-497/195 expression and disease-free and relapse-free survival across the broad range of molecular BCP-ALL subtypes, including CDKN2A/CDKN2B or IKZF1 deletion. If the associations hold true, and particularly if further preclinical data support the use of targeted therapies when miR-497/195 expression is low, then miR-497/195 expression testing may well add not only to the refinement of BCP-ALL risk stratification, but also point a way to rational, personalized, biology-informed improvements in therapy.
Conflict-of-interest disclosure: The authors declare no competing financial interests.