In this issue of Blood, Park et al1 expand the list of putative driver genes in cutaneous T-cell lymphoma (CTCL). They also show that PDCD1 deletion leads to a reversal of T-cell exhaustion signatures and is associated with a worse prognosis.
CTCL is a heterogeneous group of diseases that is characterized by clonal expansion of malignant T cells primarily in the skin. Mycosis fungoides (MF) and Sézary syndrome (SS) are the classic examples of CTCL and account for ∼60% and 5%, respectively, of all CTCLs. MF is an epidermotropic primary CTCL that includes multiple clinicopathological presentations, from isolated MF lesions to erythroderma and visceral disease that often evolve over time. According to the World Health Organization classification,2 the term MF should only be used for cases that are characterized by the evolution of patches, plaques, and tumors. SS is defined by erythroderma, generalized lymphadenopathy, and neoplastic T cells in the skin, lymph nodes, and peripheral blood. The prognosis of MF and SS varies from >20 years to <1 year.
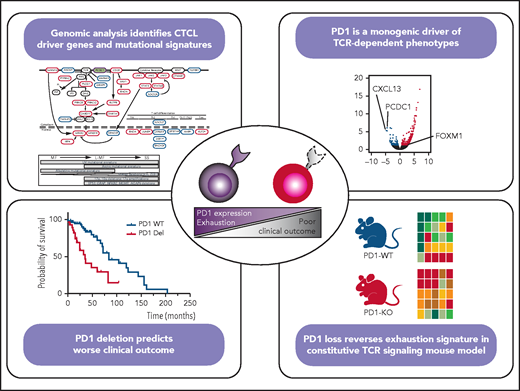
In the last few years, genome sequencing has provided important insights into the biology of these entities.3-8 Despite these advances, we do not entirely understand the causes of CTCL heterogeneity. This study by Park et al explores a group of 95 new CTCL samples that were characterized by whole-genome and whole-exome sequencing; they combined their data with previously published data from 203 samples.4 This large number of samples from diverse disease subtypes and stages allowed them to identify new putative driver genes. They found hotspot point mutations in the oncogenes NFKB1 (p.H67Y), KLF2 (p.H346Q/N/Y), and JUNB (p.A282V), as well as damaging mutations in the tumor suppressor genes FUBP1 and ANO6. They used the whole-genome sequencing data from 75 samples to identify new significant structural variants, including highly recurrent deletions in GRAP, AGAP6, ZBTB7A, and SBNO2. They also found translocations affecting the BACH2 gene in 14% of the samples. The list of new putative driver genes is dominated by genes with roles in T-cell receptor (TCR) signaling (GRAP), cytokine signaling (SNBO2), and T-cell differentiation (BACH2, KLF2, NFKB1). It is of particular interest that the investigators compared the mutation patterns of early-stage MFs, late-stage SS, and MFs that had progressed to erythroderma, lymphadenopathy, and leukemia (leukemic MF [L-MF]). They found that UV-damage signatures were enriched in all of the samples, irrespective of their diagnosis; alkylating-related signatures were restricted to early MFs, and age-related signatures were enriched, but not exclusively, in SS (see figure). They described the distribution of the structural variants, which were rare in early-stage MF cases, but 17p and 10q deletions and 17q amplification were recurrently found in leukemic forms. Although many driver mutations were present in both forms, the mutations were more prevalent in L-MF and in SS compared with skin-limited CTCL. Their comparative analyses did not include late-stage tumoral MF restricted to the skin and without blood involvement; therefore, further investigation is needed to distinguish whether this prevalence is due to their leukemic nature or to the more aggressive stages of L-MF and SS. However, the results do show genetic commonalities between MF and SS, suggesting that they are more likely to represent 2 extremes of the same spectrum than to be different diseases.
They also studied the effect of the CTCL genotype on its phenotype heterogeneity. To this end, they performed a multimodal analysis of a selected group of samples, using whole-genome sequencing data, TCR ex vivo stimulation of sorted malignant cells from SS and frozen skin MF tumors, and TCR-dependent functional analysis of proliferation and cytokine production. The investigators exposed CTCL leukemic samples to agonistic TCR stimulation and found significant heterogeneity in their proliferative capacity. Some CTCL samples showed characteristics of the so called “T-cell exhaustion” phenotype. T-cell exhaustion is defined as a dysfunctional state in which there is a failure to form functional memory T cells, with increased expression of inhibitory receptors, reduced cytokine production, weakened cell proliferation potential, and an altered transcriptional program. By combining RNA sequencing and cytometry by time-of-flight data, they found programmed cell death protein 1 (PD-1) to be the most significantly upregulated gene in nonproliferative samples. Samples with a “fully exhausted” phenotype could not proliferate after TCR stimulation and expressed higher levels of the exhausted phenotype markers, PD1 and TGT1, as well as CXCL13, than did normal CD4+ T cells. In contrast, low PD-1 expressors (unexhausted samples, with a gain-of-function phenotype) produced more effector cytokines, had an upregulated cell cycle transcription pattern, and harbored PDCD1 deletions. This led the investigators to suggest that PD-1 is the driving force giving rise to this heterogeneity (see figure).
Furthermore, the investigators found that the PDCD1 gene, which encodes PD-1, was deleted more frequently in high-proliferative samples. They validated this result in a constitutive TCR signaling murine model, confirming the PD-1–dependent differences in exhaustion phenotypes in CTCL samples. They also showed that loss of the PDCD1 gene is associated with more aggressive stages and shorter overall survival.
In summary, previous studies claimed that the biology of malignant T cells in MF and SS was distinct in MF and SS, at least in part as a result of the expression of cell surface markers consistent with skin-resident effector memory T cells and central memory T cells, respectively.9 In fact, the WHO classification described them as being closely related, but different, entities. However, molecular evidence, including that offered by Park et al, indicates that MF and SS may be 2 extremes of the same spectrum. This point is still not resolved to everybody’s satisfaction and remains open to discussion.
Genomic analysis in CTCL patients identifies new genetic drivers and mutational signatures shared by MF and SS samples (see Figures 2 and 3 in the article by Park et al that begins on page 1225). A multimodal analysis using TCR ex vivo stimulation of malignant cells and TCR-dependent functional analyses shows that PD-1 loss leads to reversal of T-cell exhaustion signatures in humans and mice and is associated with a worse prognosis (see Figures 4 and 6 in the article by Park et al). The figure was prepared by Natalia Yanguas-Casas, using Figures 2, 3G, 4G, and 6D in Park et al. KO, knockout; Tfh, T follicular helper; Th, T helper; Trm, tissue-resident memory T cell; WT, wild-type. Putative oncogenes and tumor suppressors are indicated by red and blue boxes, respectively. *Indicates genes not previously reported in CTCL.
Genomic analysis in CTCL patients identifies new genetic drivers and mutational signatures shared by MF and SS samples (see Figures 2 and 3 in the article by Park et al that begins on page 1225). A multimodal analysis using TCR ex vivo stimulation of malignant cells and TCR-dependent functional analyses shows that PD-1 loss leads to reversal of T-cell exhaustion signatures in humans and mice and is associated with a worse prognosis (see Figures 4 and 6 in the article by Park et al). The figure was prepared by Natalia Yanguas-Casas, using Figures 2, 3G, 4G, and 6D in Park et al. KO, knockout; Tfh, T follicular helper; Th, T helper; Trm, tissue-resident memory T cell; WT, wild-type. Putative oncogenes and tumor suppressors are indicated by red and blue boxes, respectively. *Indicates genes not previously reported in CTCL.
The most exciting finding is the putative role of the PD-1 gene in CTCL. Although gene mutations in CTCL commonly promote TCR-dependent proliferation, most CTCL cases show “T-cell exhaustion” characteristics. Their analyses identify PD-1 as being responsible for this: loss of PD-1 is enough to reverse this phenotype, increasing cell proliferation and prompting a worse clinical course. Previous studies reported that the neoplastic T cells in SS express PD-1 in most cases.10 The finding that the loss of PD-1 is associated with a worse prognosis is an intriguing one that requires further functional studies and validation in larger series.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal