In this issue of Blood, Shimada et al explore the molecular pathogenesis of intravascular large B-cell lymphoma (IVLBCL) and document frequent alterations of immune checkpoint–related genes that favor immune evasion. The authors identified aberrations of PD-L1, PD-L2, and other genes in plasma DNA. These findings open new opportunities for diagnosing and monitoring IVLBCL.1
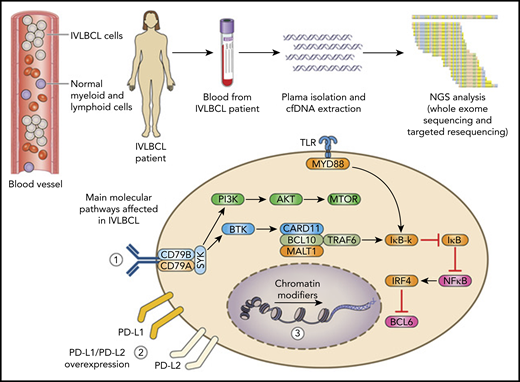
IVLBCL selectively localizes to the lumen of blood vessels where the lymphoma grows and disseminates. Using a liquid biopsy strategy that exploits whole-exome sequencing has elucidated the disease genomic landscape. The affected molecular pathways include (1) BCR/NF-κB, (2) immune checkpoint regulators (PD-L1 and PD-L2), and (3) chromatin remodeling. Molecular lesions of PD-L1 and PD-L2 induce overexpression of the corresponding protein, which highlights immune escape as a major pathogenetic feature of IVLBCL that can be targeted with immune checkpoint inhibitors.
IVLBCL selectively localizes to the lumen of blood vessels where the lymphoma grows and disseminates. Using a liquid biopsy strategy that exploits whole-exome sequencing has elucidated the disease genomic landscape. The affected molecular pathways include (1) BCR/NF-κB, (2) immune checkpoint regulators (PD-L1 and PD-L2), and (3) chromatin remodeling. Molecular lesions of PD-L1 and PD-L2 induce overexpression of the corresponding protein, which highlights immune escape as a major pathogenetic feature of IVLBCL that can be targeted with immune checkpoint inhibitors.
IVLBCL is a rare type of aggressive large B-cell lymphoma with a selective tissue tropism that causes an almost exclusive localization of tumor cells within the lumen of blood vessels, with a preference for smaller vessels and capillaries.2,3 Remarkably, the blood vessel is the site of active cell replication and also serves as vehicle for disease spread. The clinical manifestations of IVLBCL are extremely heterogeneous, and the disease is frequently regarded as a great imitator, which poses diagnostic challenges,3,4 but increasing clinical awareness of it has steadily improved recognition and diagnosis.5 A skin biopsy is generally used to reach a diagnosis in cutaneous IVLBCL and in a fraction of patients with the classical variant of the disease. The diagnostic process is more challenging in patients who do not have skin involvement because of the current lack of disease biomarkers and because sites of lymphoma may not be apparent on imaging studies.
The rarity of IVLBCL, combined with difficulties in adequate tumor sampling for large-scale molecular studies, has prevented a comprehensive understanding of its pathogenesis. Most occurrences of IVLBCL, as many as 70% to 80%, belong to the non-germinal center B-cell–like (non-GCB) category according to the Hans algorithm, and the disease immunophenotype may overlap with that of diffuse large B-cell lymphoma (DLBCL) not otherwise specified.2,3 Consistent with the non-GCB phenotype of IVLBCL, mutations of genes belonging to the BCR/TLR/NF-κB signaling pathways have been reported.6,7
The study by Shimada et al aimed to elucidate the pathogenesis of IVLBCL by using a whole-exome sequencing strategy performed mainly on the patients’ cell-free DNA (cfDNA). This strategy was based on the assumption that the intravascular nature of the disease might represent an ideal model for a liquid biopsy investigation, as is also suggested by pivotal evidence (see figure).7 This approach proved to be valuable and informative: several molecular alterations with pathogenetic relevance were unequivocally identified in the IVLBCL patients who were analyzed; the median mutation load (236.5 mutations per sample) was higher in the patients’ plasma than in the bone marrow and, importantly, cfDNA levels in the plasma of IVLBCL patients correlated with lactate dehydrogenase at diagnosis and rapidly decreased after starting therapy in responding, but not in refractory, patients.
Structural variations of the immune checkpoint–related genes PD-L1 and PD-L2 emerge as a remarkable and frequent feature of IVLBCL, occurring in almost half the patients (see figure).
These molecular alterations of PD-L1 and PD-L2 almost consistently truncate the 3′ untranslated region of the affected gene, lead to transcript stabilization and overexpression, and associate with strong immunostaining of the protein in IVLBCL cells carrying these lesions. Rearrangements of PD-L1 and PD-L2, coupled with alterations of HLA molecules, may represent an important mechanism of immune escape for IVLBCL cells that would otherwise be continuously exposed to immune effector cells circulating in the bloodstream. The genetic landscape of IVLBCL also shares mutations of several driver genes that are involved in other variants of DLBCL, in particular of the non-GCB type (see figure).8 The affected genes belong to the BCR/NF-κB signaling pathway, to chromatin modifiers and remodelers, and to genes involved in B-cell differentiation (see figure).
The genetic features of IVLBCL highlighted by Shimada et al may serve as disease biomarkers, especially since they can be detected in the patients’ cfDNA (see figure). Liquid biopsy of IVLBCL emerges as a valuable and sensitive approach for a comprehensive view of the disease genotype, because several gene alterations can be identified uniquely in the cfDNA and not in bone marrow samples. The potential relevance of liquid biopsy in B-cell lymphoma goes beyond the role of assisting and complementing diagnosis in the challenging clinical settings that are frequently encountered in patients affected by IVLBCL. The fact that liquid biopsy that exploits the tumor genetic landscape is able to predict response to treatment in other B-cell lymphomas provides an interesting model for IVLBCL.9,10 Of note, analysis of cfDNA has been incorporated into integrated models for risk prediction that, although they were designed for DLBCL and Hodgkin lymphoma, might also be adapted to IVLBCL in the future.9,10 From this standpoint, the results from the study by Shimada et al provide pivotal evidence of the potential clinical significance of cfDNA variations during treatment for IVLBCL, with a rapid decrease in cfDNA levels in patients who respond to therapy. Liquid biopsy of IVLBCL may also have potential implications in disease monitoring after achievement of response. In fact, in a clinical condition such as IVLBCL, in which imaging studies have limitations for the early detection of relapse, liquid biopsy may provide an important advantage for noninvasive molecular monitoring over time.
These advances in the molecular pathogenesis of IVLBCL represent a significant step forward in the understanding of this rare and peculiar disease entity. In parallel, these molecular notions provide a strong rationale for designing translational and clinical studies dedicated to the refinement of our current management of IVLBCL in terms of improving diagnostic recognition and prognostic stratification, designing treatment strategies, and monitoring the disease at the molecular level. For example, the detailed knowledge of the genetic landscape of IVLBCL helped generate a panel of biomarkers that can be tested in large retrospective and prospective cohorts in studies aimed at defining a diagnostic and prognostic stratification of IVLBCL heterogeneity and at designing a precision medicine algorithm for the disease. In addition, the high prevalence of PD-L1 and PD-L2 alterations associated with PD-L1 and PD-L2 overexpression on the lymphoma cell population points to evasion from antitumor immunity as an important pathogenetic feature of IVLBCL that might be targeted and reverted, at least in part, with the therapeutic use of immune checkpoint inhibitors that are already available for clinical use. A liquid biopsy approach based on the IVLBCL genotype, similar to that used in other lymphomas,10 might provide a useful tool for monitoring disease responsiveness to treatment in real time and for predicting early relapse during the clinical follow-up.
Conflict-of-interest disclosure: A.P. declares consulting fees from Ariad, Sanofi, and Takeda, all outside the scope of this work; G.G. declares consulting and lecturing fees from Janssen, Abbvie, and Astra-Zeneca, all outside the scope of this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal