Substantial progress in the treatment of Multiple Myeloma (MM) extends survival for many patients (Pts), though most Pts eventually relapse and become therapy refractory. Patients with induction resistant multiple myeloma (IRMM), either primary refractory or early (≤18 months) relapse, have a particularly compromised survival. New treatment strategies and molecular biomarkers for patient stratification and effective clinical care are needed.
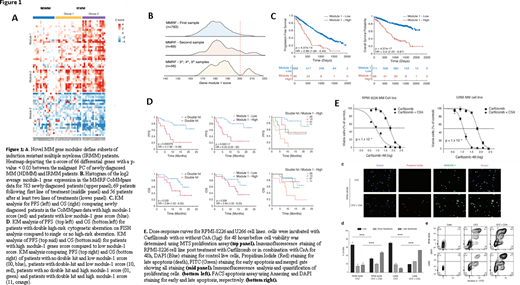
We previously reported outcomes of KYDAR (NCT04065789) single-arm prospective clinical trial, in which pts primary resistant to a bortezomib-based induction achieved high rates of durable responses when treated with carfilzomib/daratumumab/lenalidomide/dexamethasone (Cohen YC et al. Blood (2019) 134 (Suppl 1): 982). We applied comprehensive single cell RNA-seq analysis of plasma cells (PCs) obtained from longitudinal bone marrow aspirate samples, taken from KYDAR participants (n=34), compared to newly diagnosed MM Pts (n=15) and to healthy controls (n=11). We discovered a novel MM resistance signature differentially expressed between IRMM and newly diagnosed MM groups. This "gene module is enriched for several pathways that were perturbed in the IRMM Pts, including mitochondrial stress genes, the ER and UPR pathway, and the proteasome machinery. Furthermore, differential gene expression analysis between KYDAR responders and non-responders unveil potentially druggable escape mechanisms. These include upregulation of genes associated with immune regulation, proteasome, apoptotic and ER-stress pathways, e.g. Cyclophilin A (PPIA) creating an elaborated signature and potential target list of pathways and escape mechanisms from a highly potent quadruple therapy. This signature includes many novel genes which were not previously described in the context of MM (Fig 1A).
Here we report external validation of this novel resistance signature among 908 MM Pts in the MMRF CoMMpass dataset. We found that our genes signature expression follows a normal distribution with no apparent sub-populations in naïve patients, but when examining Pts after multiple relapses, we detected gradient increase in our signature with a clear bi-model distribution (Fig. 1B). The prevalence of high module-1 expression was 5% in newly diagnosed Pts vs 14% in Pts in 3rd or subsequent relapse (p<0.001). Survival analysis on MMRF "module 1 high" (module 1 score > 200) Pts (n = 68) compared with the rest of the population (n = 711) revealed a striking hazard-ratio of 3.9 (2.22 - 6.87) with p-value = 4.57x10-17 (Fig 1C). Module-1 was highly predictive of treatment outcome in KYDAR trials, beyond FISH cytogenetics (Fig 1D).
We hypothesized that PPIA may function as a protective resistance gene in MM malignant cells, by accelerating protein folding pathways and reducing stress associated to proteasome inhibitors. In order to test whether PPIA is merely a marker for highly resistant patients or has a causal role in MM resistance to proteasome inhibitors, we used Cyclosporine A (CsA), a known inhibitor of PPIA, in a series of in vitro experiments, to explore it's potential synergy with carfilzomib, a proteasome inhibitor, on RPMI-8226 and U266B MM cell lines, expressing high levels of PPIA. Using MTS proliferation assay, we found that the combined CsA and carfilzomib therapy was significantly more effective than carfilzomib alone. Apoptosis as measured by Propidium Iodide, DAPI and Annexin V FITC staining, was dramatically increased in the combination therapy setting compared to carfilzomib or CsA monotherapy (Fig 1E).
In summary, our study defines a roadmap for combining single cell RNA-seq profiling with clinical trials. We reveal and externally-validate a novel transcriptional signature for therapy resistance. We show inhibition of PPIA, a potential target identified, by CsA, overcomes relative resistance of MM cell lines to carfilzomib. We anticipate that such studies will significantly improve the ability to define mechanism of action of treatment, molecularly characterize the Pts that may benefit from the treatment, and reveal potential novel targets.
Tadmor:AbbVie: Consultancy, Speakers Bureau; Janssen: Consultancy, Speakers Bureau; Takeda: Consultancy, Speakers Bureau; Sanofi: Consultancy, Speakers Bureau; Medison: Consultancy, Speakers Bureau; Neopharm: Consultancy, Speakers Bureau; 6. Novartis Israel Ltd., a company wholly owned by Novartis Pharma AG: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal