Background
Over the last decade, the advent of next-generation sequencing (NGS) has dramatically improved our understanding of the genomic landscape that characterize acute myeloid leukemia at time of diagnosis, leading to the identification of new prognostic subsets and therapeutic targets. Still, few studies have addressed how this landscape changes upon treatment, and in particular after allogeneic hematopoietic cell transplantation (allo-HCT). Moreover, the links between the mutational profile of AML and mechanisms of post-transplantation immune evasion remain to date largely unknown.
Methods
Taking advantage of the HaloPlex High Sensitivity (HS) technology, which allows a more precise definition of mutations and clonal architecture through the use of unique molecular identifiers, we designed and tested a NGS panel covering genes and miRNAs known to be involved in the pathogenesis of myeloid malignancies (n=112), in the DNA damage response (n=50) or in immune-related processes (n=30). By using an Illumina HiSeq2500n instrument,we sequenced a total of 200 AML samples collected from 131 patients, including 84 diagnoses, 45 relapses after chemotherapy (CT) and 71 relapses after allo-HCT, 31 of which characterized by genomic loss of HLA (Vago et al, N Engl J Med, 2009). Variant calling was performed using a pipeline based on the FreeBayes algorithm.
Results
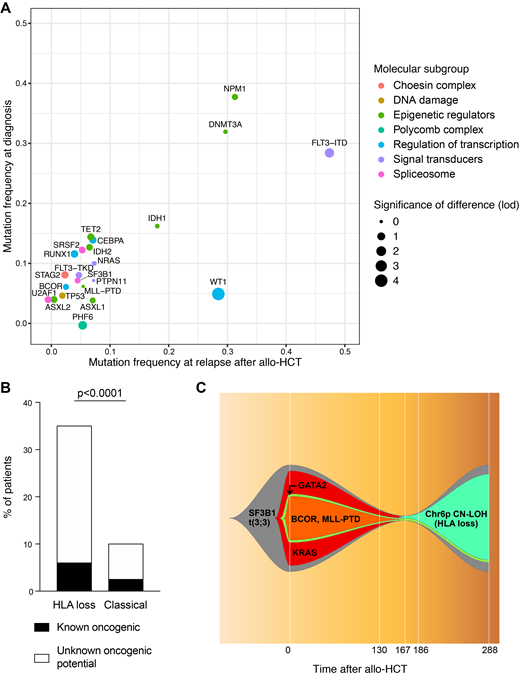
Sequencing coverage was uniform across all target amplicons, with a 855x average depth-of-sequencing, and a mean of 122 unique barcodes for each region. Among the 84 diagnosis samples we identified 253 oncogenic mutations, with a median of 3 mutations per patient (range 0-8). Both the frequency and the prognostic value of mutations detected at diagnosis were in line with the largest published dataset on AML genomics (Papaemmanuil et al, N Engl J Med, 2016; R2= 0.88, p<0.0001). The overall burden of oncogenic mutations did not change significantly neither at relapses after CT nor after allo-HCT. Analyzing the profile of known oncogenic drivers of diagnoses and of the two relapse cohorts we documented high similarity between the three time-points, with significant differences limited to FLT3-ITD (present in 29% patients at diagnosis vs 49% and 48% at relapses after CT and allo-HSCT, p=0.02 and p=0.01 respectively) and WT1 mutations (5% vs 20% and 28%, p=0.006 and p<0.0001 respectively; Figure 1A). Also when comparing post-transplantation HLA loss relapses with their "classical" counterparts we could find no significant difference in the frequency or co-occurrence pattern of known AML driver mutations. Still, in HLA loss relapses we documented a significantly higher frequency of variants of unknown oncogenic potential befalling in genes implicated in the DNA damage response, including those belonging to the Fanconi Anemia pathway (35% of mutations in HLA loss vs 10% in classical relapses, p<0.0001; Figure 1B). This intriguing observation suggests that, in these cases, double strand breaks might be more frequently repaired through homologous recombination, and that this in turn might result in a higher likelihood of copy-neutral loss of heterozygosity events, the molecular mechanism at the basis of HLA loss. Finally, thanks to our panel, we were able to track possible routes of leukemic clonal evolution between diagnosis and post-transplantation relapse: interestingly, in several cases emergence of an HLA loss subclone out-competed clones carrying known oncogenic drivers, suggesting that after allo-HCT the immune privilege might outperform the proliferative advantage (Figure 1C).
Discussion
Taken together, our data highlight a complex interplay between the genomic landscape of AML and the environmental selective pressure represented by the graft-versus-leukemia effect. Whereas in fact some genomic alterations that carried little consequence before transplant become able in this setting to promote specific patterns of immune escape, others, that had driven AML biology before, turn out to be dispensable after allo-HCT. Understanding and tracking these evolutionary dynamics may provide new tools and targets to treat, or even prevent, AML post-transplantation relapses.
Sala:Gilead Company: Honoraria. Gentner:Genenta Science: Consultancy, Equity Ownership, Research Funding. Vago:GenDx: Research Funding; Moderna Therapeutics: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal