Introduction: Acute Myeloid Leukemia (AML) is a genetically heterogeneous disease, associated with mutations in a well-established set of genes that affect myeloid cell differentiation and proliferation. While treatment options were limited for many decades, many new pharmacological treatments in the form of small molecule inhibitors have been developed in recent years. In order to be effective, many of these inhibitors require the presence of specific biomarkers that must be detected with appropriate genetic screening assays. Genetic studies may also inform conventional care plans. The cost and time of genetic testing can be limiting factor, delaying the start of treatment and often requiring referral to larger regional medical center. Such delays can result in increased morbidities and mortality in this patient population, making faster and more accessible genetic testing highly relevant.
Methods: We have developed a rapid single-molecule sequencing-based assay which requires minimal laboratory equipment, while providing same-day results. We utilized amplification-based library preparation to generate sequencing libraries for the Oxford Nanopore Technologies MinION instrument. We have combined rapid library preparation, real-time single-molecule sequencing, and novel computational analysis algorithms to rapidly screen patient blood samples for the presence of actionable biomarkers. In this study, we analyzed samples from 48 AML patients with known mutational statuses to evaluate the performance of our approach. For our proof of principle assay, we included seven molecular targets (six hotspot mutations; one duplication) in five AML-associated genes (DNMT3A, FLT3, IDH1, IDH2, and JAK2). We compared the accuracy of our assay to current gold standard approaches which include Illumina-based short-read sequencing and capillary electrophoresis.
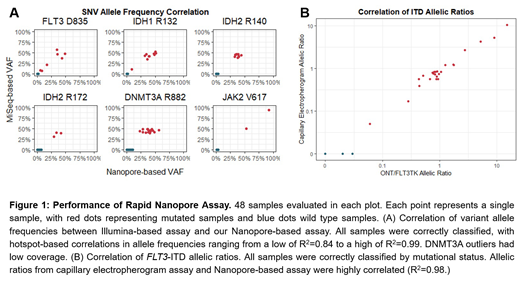
Results and Discussion: Our assay exhibited equal or superior accuracy compared to the current gold standard approaches, while providing results significantly faster (in less than 6 hours). Each of the 48 samples were correctly classified by mutational status, with the detected variant allele frequency exhibiting high correlation (R2 for mutational hotspots ranging from a low of 0.84 to a high of 0.99) between our Nanopore-based assay and the control Illumina-based assay. Our custom FLT3-ITD detection algorithm correctly identified 24/24 samples as ITD positive, with insertion lengths matching expected peak lengths from capillary electropherogram testing and highly concordant allelic ratios. In contrast, an assay using Illumina sequencing data correctly detected only 6/24 ITD positive samples. Therefore, we conclude that our assay provides superior variant detection accuracy, significantly shorter time to results (4-6 hours instead of days), and at an expected cost of below $250/sample when utilizing single-sample Flongle flow cells. Critically, this procedure is accessible beyond tertiary care academic medical centers and could be deployed at the point-of-care in smaller hospitals, physician offices, and resource-constrained settings worldwide.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal