Introduction:
Several studies have investigated the utility of digital droplet polymerase chain reaction (ddPCR) in the detection of p210 BCR-ABL fusion transcripts in chronic myeloid leukemia (CML), and have suggested greater sensitivity compared to real-time quantitative PCR (RT-qPCR). There is limited data regarding the utility and sensitivity of ddPCR in patients with chronic phase CML (CP-CML) who are candidates for tyrosine kinase inhibitor (TKI) discontinuation. We aimed to investigate the capability of ddPCR to detect p210 BCR-ABL transcript levels in CP-CML patients and compare the sensitivity to RT-qPCR.
Methods:
We analyzed 103 different peripheral blood (n=98) and bone marrow (n=5) samples from 36 patients with CP-CML treated at Moffitt Cancer Center between 2013 and 2018. Samples were from patients who either met clinical criteria for TKI discontinuation or were enrolled in a phase I dose-escalation study evaluating combination therapy with nilotinib plus ruxolitinib who had at least a complete cytogenetic response (CCyR) to TKI therapy with detectable BCR-ABL transcripts by RT-PCR at the time of enrollment(1, 2). BCR-ABL levels were assessed by RT-qPCR at the time of collection with level of transcript reported as % International Standard (%IS). Samples collected from 2013 to 2014 were reported as p210 to ABL and converted to %IS per lab-specific conversion factor. Stored purified RNA samples were obtained retrospectively and ddPCR was performed using the QX-200 Droplet Digital PCR System and QXDx BCR-ABL %IS Kit (Bio-Rad). Droplets were analyzed by QXDx BCR-ABL reporter software to determine p210 transcript level (%IS). Analysis for RNA degradation of stored samples was performed via High Sensitivity RNA ScreenTape (Agilent). Samples that did not pass quality check upon performing ddPCR were excluded from further analysis. Correlation coefficient was calculated with p-value of <0.05 being significant.
Results:
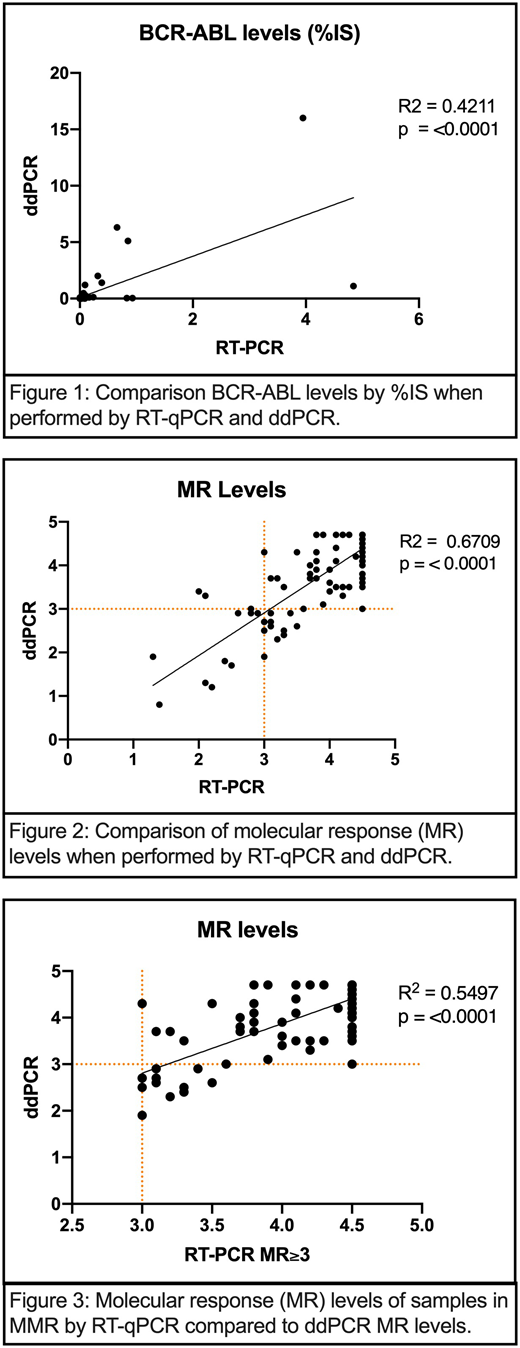
Of the 36 patients, 28 were in the TKI discontinuation cohort and 10 were enrolled in the clinical trial of nilotinib plus ruxolitinib. Two patients who participated in the clinical trial subsequently also met criteria for TKI discontinuation. In the TKI discontinuation cohort, 46% of patients (n=13) lost major molecular response (MMR) with a median time to loss of MMR of 88 days. We observed a statistically significant correlation between RT-qPCR and ddPCR levels when comparing %IS (correlation coefficient R2=0.42; p<0.0001) as well as calculated molecular response (MR) logarithm values of all available samples (R2=0.67; p <0.0001) (Figure 1). There was a weaker correlation between patients determined to have MMR by RT-qPCR (MR≥3.0) compared to ddPCR (R2=0.55; p <0.0001) (Figure 2). A total of 91 different samples were consistent with MMR when tested by RT-PCR. Of these samples, 85.7% (n=78) were consistent with MMR when tested by ddPCR and the remaining 14.3% (n=13) appeared to have higher detectable levels of p210 transcripts (MR<3.0). When investigating deeper molecular responses, 62 samples were consistent with MR≥4.0 by RT-PCR, and 20.9% (n=13) of these samples did not achieve a deep molecular response (MR<4.0) when tested by ddPCR. Of note, 5 representative samples were also evaluated for potential RNA degradation and 3 were noted to have significant degradation present.
Conclusion:
Compared to RT-PCR, ddPCR represents a viable method for detection of p210 transcripts for patients with CP-CML. The two methods yield similar efficacy when detecting varying levels of molecular response, however further research is warranted for the ability to detect deeper molecular responses for patients in MMR. Our study is limited by inherent retrospective bias and quality of RNA in stored samples.
1. National Comprehensive Cancer Network. Chronic Myeloid Leukemia (Version 1.2019).
2. Sweet K, Hazlehurst L, Sahakian E, Powers J, Nodzon L, Kayali F, et al. A phase I clinical trial of ruxolitinib in combination with nilotinib in chronic myeloid leukemia patients with molecular evidence of disease. Leuk Res. 2018;74:89-96.
Talati:Jazz Pharmaceuticals: Honoraria, Speakers Bureau; Daiichi-Sankyo: Honoraria; Astellas: Honoraria, Speakers Bureau; Pfizer: Honoraria; Celgene: Honoraria; Agios: Honoraria. Shelton:Bio-Rad: Employment. Nodzon:Pharmacyclics: Consultancy; Abbvie: Other: Speaker Fees; Genentech: Consultancy, Other: Speaker Fees; Pfizer: Consultancy. Sweet:Stemline: Consultancy; Pfizer: Consultancy; Abbvie: Membership on an entity's Board of Directors or advisory committees; Celgene: Speakers Bureau; Bristol Myers Squibb: Membership on an entity's Board of Directors or advisory committees; Astellas: Membership on an entity's Board of Directors or advisory committees; Incyte: Research Funding; Jazz: Speakers Bureau; Novartis: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Agios: Membership on an entity's Board of Directors or advisory committees. Pinilla Ibarz:Novartis: Consultancy; Abbvie: Consultancy, Speakers Bureau; Takeda: Consultancy, Speakers Bureau; Bristol-Myers Squibb: Consultancy; Janssen: Consultancy, Speakers Bureau; Bayer: Speakers Bureau; TG Therapeutics: Consultancy; Teva: Consultancy; Sanofi: Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal