Background: The group of rare hereditary anemias includes a large variety of intrinsic defects of the red blood cell, as well as erythropoiesis. They include hemolytic anemias (e.g. enzyme deficiencies), hemoglobinopathies, hypoplastic anemias (e.g. Diamond-Blackfan Anemia, DBA), and dyserythropoietic anemias. As a result of the rapid developments in genetic testing and the subsequent increased knowledge of molecular defects underlying hereditary anemias, our understanding of the pathophysiology of rare anemias has increased during the last decade. However, in a substantial number of patients, the clinical phenotype does not fit classical criteria of a disease, response to therapy is less than expected, or a molecular defect cannot be found. In addition, in patients with well-described molecular defects, there is often no clear genotype-phenotype correlation. In order to better understand the underlying pathophysiological mechanisms driving ineffective erythropoiesis in patients and to improve their classification and clinical evaluation, novel functional tests are needed. Metabolomics is the large-scale, unbiased study of metabolites and their interactions within a biological system, directly reflecting the underlying biochemical activity and state of cells. Metabolomics can be used to identify novel disease biomarkers, study deregulated cellular pathways, and to determine the cellular responses to therapeutic interventions. In this study we demonstrate that dried blood spots (DBS) can be used as a minimal invasive and validated technical approach to perform large scale metabolomics in a variety of rare hereditary anemias.
Methods:
DBS samples from >100 patients suffering from a variety of rare anemiaswere collected during regular hospital visits. Quantification of metabolites was performed by direct infusion high resolution mass spectrometry (DI-HRMS) followed by an untargetedmetabolomics pipeline. For annotation, the Human Metabolome Database (HMDB) was used. Results were compared with DBS samples of 70 healthy adult controls and 35 pediatric patients negatively screened for metabolic diseases
Results:
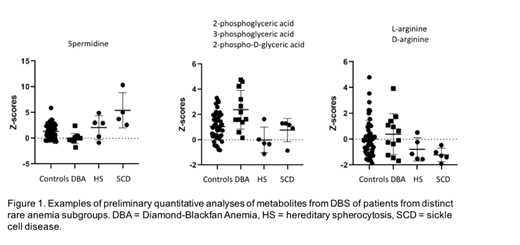
For each patient sample, Z-scores were calculated for all mass peaks annotated with metabolites (HMDB, 3930). Mass peak, intensity and corresponding Z-scores were compared with two distinct groups of controls (∆Z-scores): pediatric patients who were screened for metabolic diseases but were found negative, and healthy adult controls. For data interpretation, two strategies were used. First, by untargeted statistical analysis in Metabo-analyst, we identified metabolites (and/or isomers) that showed either increased or decreased intensity. For the second strategy we specifically focused on red blood cell metabolic pathways, including glycolysis, the pentose phosphate pathway, ascorbate and glutathione metabolism, arginine and polyamine metabolism, and erythrocyte membrane turnover and transport. We corrected for a potential hematocrit effect and performed subgroup analyses correcting for reticulocyte counts. Our preliminary data indicate potential biomarkers for distinct disease entities, including altered polyamine metabolism (DBA, SCD), glycolysis (DBA, HS), and aberrant arginine metabolism (SCD) (Figure 1).
Further in-depth pathway analyses, and targeted validation of biomarker profiles are currently being performed.
Conclusion:
Untargeted metabolomics using dried blood spots provides a novel functional tool to identify disease biomarkers and common and distinct deregulated cellular pathways. This will improve diagnostic evaluation and clinical management of patients with rare hereditary anemias, contribute to a better understanding of disease pathophysiology, and aid in the development of therapeutic strategies.
van Beers:Agios Pharmaceuticals, Inc.: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Research Funding; Pfizer: Research Funding; RR Mechatronics: Research Funding. van Wijk:RR Mechatronics: Research Funding; Agios Pharmaceuticals: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal