Multiple Myeloma (MM) is a plasma cells malignancy with number of recent therapeutic options that has improved outcomes with median survival now stretching beyond 8 years. There has been an intense search to identify genomic and laboratory correlates of outcome for high risk patients. However, a subgroup of patients have a long survival but genomic segmentation of this important group which can be potentially cured has not been identified. We here described first such attempt at identifying a subset of patients with long survival.
We have analyzed data from 205 newly-diagnosed uniformly-treated MM patients using both deep whole genome (WGS) (80x) and whole transcriptome sequencing (RNAseq). Median number of SNVs, small insertion and deletion per megabase were 2.18 [0.49 - 14.52], 0.077 [0.024 - 0.17] and 0.12 [0.03 - 0.26] respectively. MM subgroups defined by FISH or copy number alterations (CNAs) had significantly different mutational load (Kruskal-Wallis p-value = 0.003). In general, Hyperdiploid MM (HMM) patients had lower SNV load compared to other subgroups and t(14;16) had the highest mutational load per megabase (Dunn test FDR < 0.25). These subgroups also showed significant differences for mutational process utilization. Mutational processes associated spontaneous deamination of 5-methylcytosine to thymine was significantly high in HMM, APOBEC activity was very strong in t(14;16) MM and DNA repair was significantly different in t(6;14), del17p and t(11;14). Importantly, the mutations associated with clonal cell population and thus in early phases of MM cell progression were driven by spontaneous or enzymatic deamination (Signature 1), somatic hypermutation in lymphoid cells (Signature 9) and Signature 17 (ANOVA p value < 1e-16) and surprisingly APOBEC activity was constant among the clones (ANOVA p value > 0.05). However, mutations associated with homologous recombination (HR) and nucleotide excision repair (NER) activity were significantly enriched in the subclonal mutations (ANOVA p value < 1e-16) suggesting their role in later stages of MM progression.
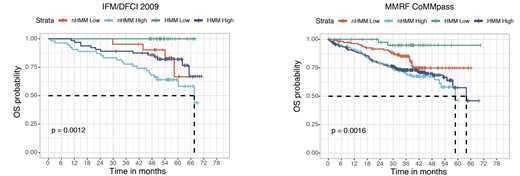
Based on this information, we next calculated the homologous recombination repair defects using copy number alterations from WGS data. We identified that HR score generated using WGS copy number information predicted outcome in newly diagnosed myeloma. Further analysis of this data identified that newly diagnosed MM patients with hyperdiploid MM (HMM) and no HR- repair deficiency have superior outcome with 6 year overall survival (OS) probability for this of 100% in IFM/DFCI 2009 cohort (Logrank test p value = 0.0012). We next validated this finding using MMRF CoMMpass dataset and we confirmed 6-yr OS probability of 95% (Logrank test p value = 0.0016) in this independent dataset (Figure 1). We further investigated RNAseq data between these genomically defined groups and identified that chromosome and telomere maintenance pathways (FDR < 0.01) and low bone disease associated genes described by Zhan et al. (FDR = 0.004) were up regulated in low-risk group. Finally, patients who were able to maintain HR, had lower mutation rate for TP53 (0% vs 9%, FDR < 0.05), CSMD1 (0% vs 6%, FDR < 0.05), FAT3/FAT4 mutations (1.7% vs 15%, FDR < 0.05), however surprisingly higher mutation rate for NRAS (42% vs 14%, p value < 0.001). In addition, DNA damage associated processes predicted by SNVs trinucleotides were significantly lower in the low-risk group (12% vs. 20%, p value < 1e-05) providing further support to this data.
In conclusion, we report a detailed genomic profile using deep DNA and RNA sequencing and identify a genomically-defined sub group that is predicted to have a long survival. This study also identifies the role of HR in myeloma with potential for its translational application in both prognosis as well as therapy.
Richardson:Janssen: Membership on an entity's Board of Directors or advisory committees; Oncopeptides: Membership on an entity's Board of Directors or advisory committees, Research Funding; Sanofi: Membership on an entity's Board of Directors or advisory committees; Karyopharm: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Bristol-Myers Squibb: Research Funding; Takeda: Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Membership on an entity's Board of Directors or advisory committees. Moreau:Celgene: Consultancy, Honoraria; Janssen: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Takeda: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria. Thakurta:Celgene: Employment, Equity Ownership. Anderson:Sanofi-Aventis: Other: Advisory Board; Bristol-Myers Squibb: Other: Scientific Founder; Oncopep: Other: Scientific Founder; Amgen: Consultancy, Speakers Bureau; Janssen: Consultancy, Speakers Bureau; Takeda: Consultancy, Speakers Bureau; Celgene: Consultancy, Speakers Bureau. Avet-Loiseau:celgene: Consultancy, Other: travel fees, lecture fees, Research Funding; takeda: Consultancy, Other: travel fees, lecture fees, Research Funding. Munshi:Takeda: Consultancy; Oncopep: Consultancy; Adaptive: Consultancy; Amgen: Consultancy; Celgene: Consultancy; Abbvie: Consultancy; Janssen: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal