Introduction: Risk stratification in acute myeloid leukemia (AML) is continually being refined as we learn more about the molecular pathophysiology of this heterogeneous disease. European LeukemiaNet (ELN) 2017 used widely to stratify the 'genetic' risk of AML, but there are considerable challenges in its application, particularly in centres where molecular genetic testing either not available, or not sufficiently timely for clinical decision making. Up to a third of the study subjects cannot be stratified using a full cytogenetic-molecular model in real-time, real-life setting. There are few attempts to combine clinical features and genetic factors aiming to find a scoring system to improve AML survival prediction. There is only one to our knowledge (Sorror ML et al. 2017). This study has generated an Adapted Genetic Risk (AGR) assessment, and used it in combination with clinical risk parameters to create a novel scoring system which has now been validated using two independent cohorts.
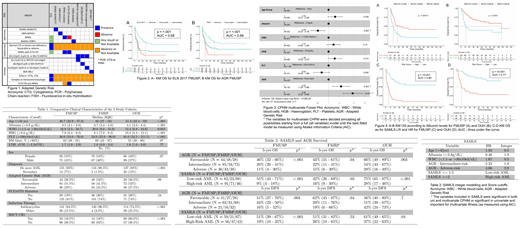
Methods: A training cohort from São Paulo (FMUSP, n = 167) of intensively treated AML patients (18-65 years) was assessed using ELN2017 genetic criteria. A comparative validation with our AGR which permits missing cytogenetic or molecular data (Figure 1) split these patients into favorable-risk (FR), intermediate-risk (IR), and adverse-risk (AR). This cohort was also used for Cox Proportional-Hazard Model (CPHM) univariate and multivariate analysis to find clinical parameters that would inform a novel Survival AML Score (SAMLS). Variables which are included in SAMLS had to be either significant in both CPHM models or significant in univariate and crucial for multivariate fitness as measured for the Akaike Information Criterion (AIC). We then applied the AGR strategy and SAMLS to 2 independent test cohorts of intensively treated adult AML patients : Riberao Preto (FMRP, n=145) and Oxford (OUH, n=157). The study was approved by the institutional review boards of the 3 participating centers. Informed consent was obtained from all patients according to the Declaration of Helsinki.
Results: Table 1 shows the clinical characteristics for all the 3 cohorts. The median follow-up (FUP) was 72.3, 44.4, and 70.5 months for FMUSP, FMRP, and OUH, respectively. The median Overall Survival (OS) was 12.4, 12.5, and 56.4 months and the 5-year OS were 29.6%, 29.7%, and 49.7% respectively.
Both ELN2017 and AGR correlated with significant differences in OS (p-value <0.001) and DFS (ELN p=0.002, AGR p=0.003) in the FMUSP cohort. The ROC c-statistic were 0.68 (ELN) and 0.66 (AGR). AGR was validated in FMRP and OUH cohorts, with statistically significant stratification of OS across the 3 risk groups in both cohorts (FMRP p=0.03, OUH p=0.003); and DFS in FMRP (p = 0.04), but not in OUH (p = 0.7)(Table 2 and Figure 2).
Multivariate CPHM model for OS in FMUSP which yielded the smallest AIC were: Age, HR 1.52; Albumin, HR 0.50; WBC, HR 1.37; Haemoglobin, HR 1.48; Platelet count, HR 0.64; and AGR-IR, HR 2.23 and AGR-AR HR, 4.66 (Figure 3). Variables which met SAMLS inclusion criteria are in Table 3.
SAMLS stratifies AML as low-risk (LR-AML) (SAMS <=1.5) or high-risk (HR AML) (SAMLS >=2) (Table 3 and Figure 4 C-D), where there is a significant difference in survival in the FMUSP training cohort (p <0.001). The 5-year OS was 55% and 9% for LR and HR AML respectively. The ROC c-statistic for OS in FMUSP using SAMLS was 0.80 (21% bigger than using AGR only). SAMLS risk stratification was validated in both FMRP and OUH cohorts for OS (FMRP p-value 0.003, OUH p-value < 0.001) and DFS (p-value 0.04 OUH only). Prediction accuracy was also increased since ROC c-statistic was 0.65 for OS in FMRP (a 10% increase), and 0.77 in OUH (a 22% increase) (Figure 4).
Conclusion: ELN2017 genetic risk score for AML was validated in a cohort from a low-medium income country (LMIC). AGR is a proposed risk evaluation that can be used for AML patients for whom fully genetic testing is not feasible (required for ELN2017) and was as accurate as ELN2017. AGR predicts survival in 3 independent cohorts, from LMIC and developed nation settings. When other biological and clinical factors are added to AGR, we were able to show SAMLS is better to predict overall survival than AGR. Importantly, serum Albumin, a simple and routine test parameter, was an independent predictor of OS for AML in all 3 cohorts. Therefore the use of SAMLS can predict early and late mortality and can impact on treatment decisions.
Quek:Celgene: Research Funding, Speakers Bureau; Agios: Research Funding. Vyas:Astellas: Speakers Bureau; Pfizer: Speakers Bureau; Abbvie: Speakers Bureau; Daiichi Sankyo: Speakers Bureau; Novartis: Research Funding, Speakers Bureau; Forty Seven, Inc.: Research Funding; Celgene: Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal