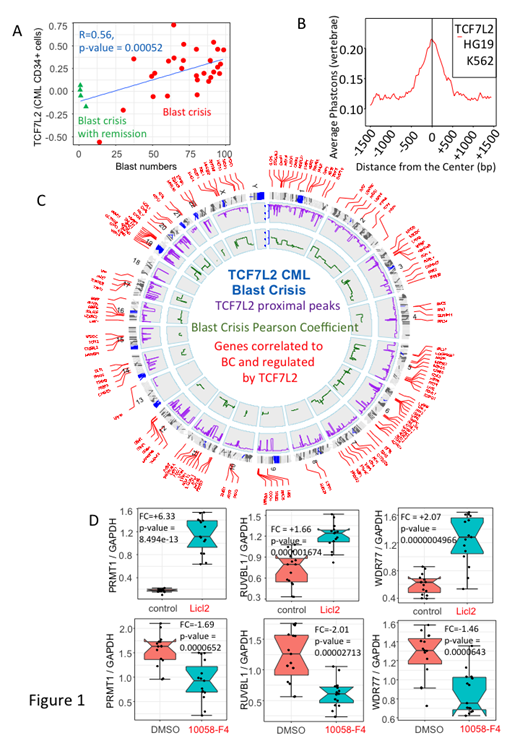
Chronic myeloid leukemia (CML) is characterized by an inherent genetic instability, which contributes to the progression of the disease towards accelerated and blast crisis (BC). The occurrence of the latter has been hampered by the use of tyrosine kinase inhibitors (TKI), which changed this natural progression, but BC still occurs in patients resistant to TKI. Several cytogenetic (major and minor routes) and genomic (TP53 mutation, p16/INK4A deletions, DNA repair abnormalities such as BRCA1, DNA-PKcs, hnRNP metabolism) events have been reported in the progression towards BC. Previous data have also suggested the involvement of embryonic stem cell program activated in BC cells such as Lin28A. In this work, we have taken advantage of the previously reported gene profiling of BC in a large cohort of patients (Radich et al. 2006) and found a correlation between blast numbers and the involvement of the Transcription Factor 7 like 2 (TCF7L2) in BC. TFC7L2 is a member of the TCF family of proteins that are known to activate WNT target genes such as Cyclin D1. TCF7L2 has been shown to be overexpressed in acute myeloid leukemia (AML) and represents a druggable target (Saenz et al Leukemia 2019). The involvement of TCF7L2 in CML-BC and its interaction with the epigenetic regulators has not been studied so far. The gene correlation study that we have performed using the blast numbers and the expression of TCF7L2 in CD34+ CML cells was found to be highly significant (Pearson test, r = 0.56, p-value=5.2e-4) (Fig.1A). TCF7L2 promoter was classed as active in K562 with ChromHMM Functional genomic analysis. K562 epigenetics peaks of TCF7L2 CHIP-seq were found principally mapped in proximal promoters (39% of the peaks, -3000pb upstream Transcription Starting Sites (TSS), Fig. 1B) and 183 unique peaks matched with promoter of 144 unique genes found to be correlated to the blast number in blood of the CML patients during BC (Fig 1C). This TCF7L2-dependent BC program was characterized to be active because promoters were also found positive for H3K27Ac and negative for H3K27Me3 histone marks, and functionally enriched with binding sites for MYC/MAX interactions (p=1.15e-6). The analysis of CHIP-sequencing of MYC revealed a significant overlapping of TCF7L2 epigenetic program with MYC (fold enrichment: 20.81, p < 2.2e-16). Surprisingly, the TCF7L2 program was found independent of RUNX1 and GATA2 transcriptional program. To determine these potential interactions, we have designed experiments in K562 cell line using the b-catenin activator Lithium Chloride (LiCL2) and the Myc/Max dimerization inhibitor 10058-F4. K562 cells were cultured in the presence of LiCL2 (10mM & 24hours) and the compound 10058-F4 (64µM & 48hours) and the expression of three epigenetic targets was analyzed by Q-RT-PCR in the presence of DMSO controls. The three targets chosen were protein arginine N-methyltransferase (PRMT1), the ATPase/Helicase RUVBL1 and the WD-repeat containing protein WDR77. As expected, after culture with LiCL2, the expression of PRMT1 was increased x 6.3 fold (p=8.49e-13) , that of RUVBL1 by x 1.66 Fold (p=1.67e-6) and that of WDR77 by x 2 fold (p=4.97e) (Fig.1D). On the contrary, the culture of K562 cells in the presence of MYC/MAX inhibitor 10058-F4, decreased the expression of 3 targets as compared to DMSO controls (x 1.6 fold for PRMT1, p=6.52e-5; x2 fold reduction for RUVBL1, p-value=2.71e-5; and x 1.4 fold for WDR77, p =0.0000643). These results show for the first time a cooperative role of TCF7L2 and MYC during blast crisis of CML and provide mechanistic insights into the interactions for the role of MYC in CML blast crisis. In addition they strengthen previous data showing a possible embryonic footprint in the blast development over the hematopoietic differentiation program during progression of the disease and provide a rationale for the pharmacological targeting of BC by the use of MYC/MAX inhibitors such as 10058-F4. Experiments are underway to evaluate the role of these factors and the MYC/MAX inhibitors in primary CML samples.
Reference : Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, Sawyers C, Shah N, Stock W, Willman CL, Friend S, Lindsey PS.(2006) :Gene Expression Changes Associated with Progression and Response in Chronic Myeloid Leukemia. Proceedings of the National Academy of Sciences of the United States of America 103 (8): 2794-99.
Turhan:Incyte: Consultancy, Honoraria; novartis: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal