Background: Lineage-specific gene transcription signatures between AML and ALL are recognized, but post-translational phenotype-specific protein expression profiles remain undefined. We hypothesized that functional proteomic patterns vary between AML and ALL and that the activity state of cells correlates with response to therapy within subgroups, complementing cytogenetic and molecular data.
Methods: Reverse phase protein arrays (RPPA) were generated using bone marrow (BM) and peripheral blood (PB) samples from newly diagnosed B-ALL (n=114), T-ALL (n=14), and AML (n=241) adult patients admitted at the MD Anderson Cancer Center. RPPA allowed simultaneous expression measurement of 229 highly validated protein antibodies including 3 Histone 3 (H3) post-translational methylation regulatory modifications; H3K4Me2, H3K4Me3 and H3K27Me3.
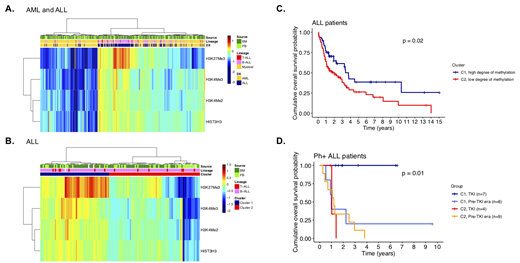
Results: Unsupervised clustering of histone modification protein expressions distinguished AML from ALL in freshly prepared lysates from BM (n=241) and PB (n=127) as well as when BM and PB samples were combined (fig. 1A). The ALL-enriched cluster was dominated by high H3K27Me3. Elevated H3K27Me3 levels were found in the BM derived leukemic blasts compared to PB blasts in ALL (P < 0.001), but not AML (P = 0.35).
Trimethylation of the repressive mark H3K27 is catalyzed by the polycomb group protein Ezh2. Oncogenic gain-of-functions of Ezh2 are seen in patients with lymphoid malignancies and others have shown that mutated Ezh2 increased H3K27Me3 in B-cells which associated with tumorigenesis. H3K27Me3 and Ezh2 antibody expressions were highly correlated in another RPPA of ALL and AML we created (R2=0.49, P < 0.001).
Profiling of methylation marks using unsupervised clustering in ALL divided patients in 2 clusters that correlated with survival (fig. 1B-C, P = 0.02). Cluster 1 (C1) with higher H3K27Me3, H3K4Me2 and H3K4Me3 was associated with better outcome. In ALL, Ph+ historically associated with poor prognosis but outcomes have improved substantially with the use of tyrosine kinase inhibitors (TKI). In our cohort, 11/26 Ph+ ALL patients were treated with TKIs and it is notable that sensitivity to TKIs correlated with cluster membership; all C1 patients (high degree of methylation) were alive after 7 years of follow-up in contrast to none of the TKI-treated Ph+ ALL patients in cluster 2 (C2, low degree of methylation) (fig. 1D, P = 0.01). Recently, TKI resistance in Ph+ ALL has been proposed to associate with smoking due to altered DNA methylation patterns caused by chemical components of cigarette smoke. Retrospectively, we identified that 2 of 11 TKI treated patients were smokers. Both had membership in C2, were resistant against TKIs and died after 1 year. Thus, 2 out of 3 resistant TKI treated Ph+ ALL were smokers compared to none of the 8 responders.
We then aimed to identify proteins that are potentially downregulated by increased expression of the repressive mark H3K27Me3. Pathway enrichment analysis of 59 significant negatively correlated proteins with H3K27Me3 revealed that these are involved in tyrosine kinase activity and resistance, including Jak/STAT and PI3K/Akt signaling pathways. If these pathways are less activated in patients with high H3K27Me3, then this can partially explain the increased sensitivity to TKIs in this subgroup. Clinically, no differences were found in age, BM and PB blast counts between TKI-treated C1 and C2 patients to provide an explanation for the higher death rate in C2.
Conclusion: ALL and AML share some pathophysiology and the identification of differences in the functional activity of cells may contribute to a better understanding of the etiology of both diseases. Here we report that high H3K27Me3 protein levels in BM and PB distinguish ALL from AML and are related to TKI sensitivity in Ph+ ALL. Histone methylation status defines a group of Ph+ ALL patients that does not benefit from the addition of TKI therapy. The idea that smoking alters the epigenetic machinery in TKI resistant Ph+ ALL has been proposed and warrants further investigation.
Fig. 1 A) Heatmap showing histone methylation levels in BM and PB from AML and ALL patients. B) Heatmap showing histone methylation levels in ALL BM and PB. Unsupervised clustering divided samples into 2 clusters. C) ALL patients in C1 survived longer than patients in C2 (P = 0.02). D) Increased long-term sensitivity for TKI therapy in C1 Ph+ ALL patients compared to C2 (100 vs. 0%, P = 0.01).
Jabbour:AbbVie: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Cyclacel LTD: Research Funding; Adaptive: Consultancy, Research Funding; Takeda: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Amgen: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal