Myeloid neoplasms (MN) are usually sporadic late-onset cancers; nevertheless, growing evidence suggests that ~5% of the cases could emerge as a consequence of inherited predisposition. Indeed, the 2016 revision of the World Health Organization (WHO) includes hereditary myeloid malignancies (HMMs) as a separate disease entity. Distinguishing somatic from germline variants is of vital importance, in order to establish an appropriate individualized management and counsel the patients and their relatives. Germline tissues are required to discriminate HMMs alterations, but they are not always available for testing at the time of diagnosis; therefore additional strategies should be put in place in order to identify variants potentially conferring genetic predisposition to MN.
The aim of the present work is to develop a tool to identify HMMs cases from tumour-only sequencing data. To this end, we reviewed tumor-only NGS reports from 299 cases of patients with myeloid malignancies sequenced with a custom Pan Myeloid Panel (PMP) that targets 48 genes, of which 21 are described in literature as HMM-associated genes. All patients signed a written informed consent form for genetic testing, research and tissue banking provided by the Biobank of the University of Navarra (UN) and were approved by the Ethical and Scientific Committees of the UN.
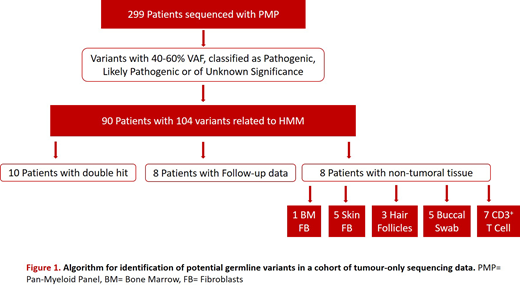
Variant calling files (VCF) of the 299 cases were run through an algorithm aiming at identifying those variants suggestive of being of germline nature (Figure 1). We considered as indicative of potential inherited origin, variants detected in bone marrow samples at a ~50% VAF classified as pathogenic, likely pathogenic or of unknown significance. This first filter yielded 90 "likely HMMs" patients harboring a total of 104 suspicious of being germline variants affecting at least one of the 21 HMM-associated genes. Our algorithm included three additional filters: mutational patterns suggestive of germline nature of the variants, analysis of variant allele frequency (VAF) on follow up data, and sequencing of non-myeloid tissues (Figure 1).
Firstly, we found 10 "likely HMMs" patients presenting double hit mutations in one of three genes: DDX41 (n=7), CEBPA (n=2) and RUNX1 (n=1). Additionally, there was available follow up data for 8 cases included in our cohort, harboring 11 suspicious variants. From them, VAFs in 4 variants affecting ASXL1, CBL, IKZF1 and GATA2 genes, drastically changed over time, indicating that those variants were somatic, whereas the other 7 variants affecting ASXL1 (n=2), DDX41 (n=2), IKZF1 (n=1), and RUNX1 (n=1) genes suspected to be germline maintained VAF ~50% with evident change of the accompanying variants over time. Finally, we collected non-myeloid tissue samples from 8 patients, harboring a total of 9 variants in ASXL1 (n=2), DDX41 (n=1), GATA2 (n=1), IKZF1 (n=1), NF1 (n=3), and SH2B3 (n=1) genes. Sequencing data confirmed germline nature of 6 of the 9 tested variants; 3 of the 6 germline variants have previously been shown to be linked to MN (DDX41 p.Asp140Glyfs*2 in UPN1, ASXL1 p.Gly967del in UPN11, and NF1 p.Met992del in UPN19). Of note, two of these bona-fide pathogenic germline mutations had been previously highlighted as potential germline variants when applying our criteria of mutational patterns and/or follow up data indicative of germline nature, in so showing the effectiveness of our algorithm (DDX41 in UPN1, and ASXL1 in UPN11).
Regarding the usefulness of the different tissues, we found that skin fibroblasts, hair follicles and CD3+ T cells helped discriminating the nature of the variants; on the contrary, DNA from hair follicles showed poor DIN values and scarce DNA concentration. Similarly, DNA isolated from buccal swab showed in general poor quality metrics, and high level of contamination with tumour DNA in one case, proving it unsuitable for discrimination of the nature of the variants in some instances.
Our data supports the importance of considering variants found upon tumor-only sequencing as potentially of germline origin, and we offer a pipeline to define the nature of the variants. We hope to provide insights for genetic laboratories facing this relatively new challenge of discriminating somatic from germline variants in tumor sequencing data.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal