Background
Acute myeloid leukemia (AML) is a clinically and molecularly heterogeneous disease with poor survival. Recurrent genetic aberrations, such as chromosomal rearrangements and gene mutations, are currently used for patient classification and prognosis, and form the basis of our current understanding of pathogenic mechanisms. However, these markers incompletely predict disease behavior and outcomes. Alterations in DNA methylation patterns are a major hallmark of cancer and recent studies have demonstrated differential global DNA methylation patterns among AML patients. Here we sought to define a novel approach to classify AML patients using genome-wide DNA methylation patterns and to uncover the biological basis and features associated with these epigenetic patterns by integrating mutation, gene expression, and functional data.
Methods
We analyzed genome-wide DNA methylation data from Illumina arrays on 649 AML cases combined from the Beat AML consortium housed at the Oregon Health & Science University (OHSU) (n=226), The Ohio State University (n=27), TCGA (n=190), and other published studies (n=206). Small molecule inhibitor response data was obtained from the Beat AML consortium. RNA sequencing and survival analyses were performed by Kaplan-Meier analysis in the TCGA and OHSU cohorts.
Results
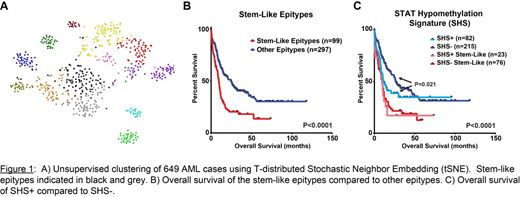
K-mediods based clustering on the 500 most-variable CpGs identified 13 distinct subtypes, we termed AML 'epitypes' (Fig. 1A). Comparative analyses incorporating normal hematopoietic cells revealed that AML epitypes retain unique combinations of biological features including the degree of myeloid development, chromatin landscapes, transcription factor binding and gene expression patterns/pathways. Eleven of 13 epitypes were associated with somatic genetic aberrations, i.e. mutations (or combinations thereof) in NPM1, IDH1/2, TET2, DNMT3A, CEBPA, and chromosomal rearrangements t(8;21), t(15;17) and inv(16) accounted for the majority of patients within each respective epitype. The remaining two epitypes were not associated with dominant genetic events, rather they clustered closely with normal CD34+ hematopoietic stem and progenitor cells. These 'stem-like' epigenetic patterns accounted for 27% of patients and showed limited correlation to other leukemic stem cell gene expression signatures. Stem-like epitypes demonstrated broad resistance to a range of small molecule inhibitors. Differential gene expression analysis between normal cells and epitypes revealed enrichment of the JAK-STAT pathway along with other inflammatory pathways, such as interleukin and interferon signaling, selectively in the stem-like epitypes. Patients with stem-like epitypes exhibited decreased overall survival relative to others independent of age (P<0.0001; Fig. 1B).
We next investigated the impact of STAT pathway activation on AML epigenetic patterns. FLT3 mutations are known to selectively activate the STAT pathway in AML and are associated with inferior outcomes. Controlling for the distribution of FLT3 mutations across epitypes, we uncovered a hypomethylation signature enriched in STAT transcription factor binding and FLT3 mutations which we have termed the 'STAT hypomethylation signature' (SHS). SHS-positivity was not restricted to FLT3 mutations, as 34% of SHS+ AMLs lacked FLT3 mutations and, conversely, SHS was absent in 24% of FLT3 mutated patients. SHS-positivity did not impact outcome in the stem-like epitypes; however, within non-stem-like epitypes, SHS+ patients displayed significantly poorer outcomes (P=0.021; Fig 1C).
Conclusion
Applying our integrative approach of using global DNA methylation profiles to subclassify AML patients, we have uncovered new insights into potential pathogenic mechanisms by which genetic and epigenetic aberrations may contribute to discrete avenues of AML development. Distinct global DNA methylation patterns are commonly driven by genetic aberrations, while others were associated with stem cell-like features and inflammatory pathways. The identification of JAK/STAT pathway upregulation in the stem-like epitype and a subset of non-stem-like samples highlights that nearly half of AML patients exhibit evidence of JAK/STAT activation and experience poor outcomes.
Druker:Dana-Farber Cancer Institute (antibody royalty): Patents & Royalties: #2524, antibody royalty; Bristol-Myers Squibb: Patents & Royalties, Research Funding; Pfizer: Other: PI or co-investigator on clinical trial(s) funded via contract with OHSU., Research Funding; Bristol-Myers Squibb: Other: PI or co-investigator on clinical trial(s) funded via contract with OHSU., Research Funding; Merck & Co: Patents & Royalties: Dana-Farber Cancer Institute license #2063, Monoclonal antiphosphotyrosine antibody 4G10, exclusive commercial license to Merck & Co; Celgene: Consultancy; Vivid Biosciences: Membership on an entity's Board of Directors or advisory committees, Other: Stock options; The RUNX1 Research Program: Membership on an entity's Board of Directors or advisory committees; Patient True Talk: Consultancy; GRAIL: Equity Ownership, Other: former member of Scientific Advisory Board; Beat AML LLC: Other: Service on joint steering committee; Cepheid: Consultancy, Honoraria; Burroughs Wellcome Fund: Membership on an entity's Board of Directors or advisory committees; Blueprint Medicines: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Beta Cat: Membership on an entity's Board of Directors or advisory committees, Other: Stock options; Aptose Biosciences: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Amgen: Equity Ownership, Membership on an entity's Board of Directors or advisory committees; ALLCRON: Membership on an entity's Board of Directors or advisory committees; ICON: Other: Scientific Founder of Molecular MD, which was acquired by ICON in Feb. 2019; Gilead Sciences: Other: former member of Scientific Advisory Board; CureOne: Membership on an entity's Board of Directors or advisory committees; Pfizer: Research Funding; Aileron Therapeutics: #2573, Constructs and cell lines harboring various mutations in TNK2 and PTPN11, licensing fees , Membership on an entity's Board of Directors or advisory committees; Monojul: Other: former consultant; Novartis: Other: PI or co-investigator on clinical trial(s) funded via contract with OHSU., Patents & Royalties: Patent 6958335, Treatment of Gastrointestinal Stromal Tumors, exclusively licensed to Novartis, Research Funding; OHSU (licensing fees): Patents & Royalties: #2573, Constructs and cell lines harboring various mutations in TNK2 and PTPN11, licensing fees . Tyner:Petra: Research Funding; Janssen: Research Funding; Aptose: Research Funding; Constellation: Research Funding; Genentech: Research Funding; Takeda: Research Funding; Syros: Research Funding; Array: Research Funding; AstraZeneca: Research Funding; Array: Research Funding; Incyte: Research Funding; Gilead: Research Funding; Janssen: Research Funding; Takeda: Research Funding; Seattle Genetics: Research Funding; AstraZeneca: Research Funding; Syros: Research Funding; Seattle Genetics: Research Funding; Constellation: Research Funding; Aptose: Research Funding; Incyte: Research Funding; Agios: Research Funding; Petra: Research Funding; Gilead: Research Funding; Genentech: Research Funding; Agios: Research Funding. Byrd:Acerta: Research Funding; Novartis: Other: Travel Expenses, Speakers Bureau; Genentech: Research Funding; Pharmacyclics LLC, an AbbVie Company: Other: Travel Expenses, Research Funding, Speakers Bureau; Janssen: Consultancy, Other: Travel Expenses, Research Funding, Speakers Bureau; Janssen: Consultancy, Other: Travel Expenses, Research Funding, Speakers Bureau; Novartis: Other: Travel Expenses, Speakers Bureau; Gilead: Other: Travel Expenses, Research Funding, Speakers Bureau; Gilead: Other: Travel Expenses, Research Funding, Speakers Bureau; Novartis: Other: Travel Expenses, Speakers Bureau; Ohio State University: Patents & Royalties: OSU-2S; Ohio State University: Patents & Royalties: OSU-2S; BeiGene: Research Funding; Acerta: Research Funding; BeiGene: Research Funding; Pharmacyclics LLC, an AbbVie Company: Other: Travel Expenses, Research Funding, Speakers Bureau; Genentech: Research Funding; Gilead: Other: Travel Expenses, Research Funding, Speakers Bureau; TG Therapeutics: Other: Travel Expenses, Research Funding, Speakers Bureau; TG Therapeutics: Other: Travel Expenses, Research Funding, Speakers Bureau; Pharmacyclics LLC, an AbbVie Company: Other: Travel Expenses, Research Funding, Speakers Bureau; Acerta: Research Funding; Ohio State University: Patents & Royalties: OSU-2S; BeiGene: Research Funding; Genentech: Research Funding; Janssen: Consultancy, Other: Travel Expenses, Research Funding, Speakers Bureau; TG Therapeutics: Other: Travel Expenses, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal