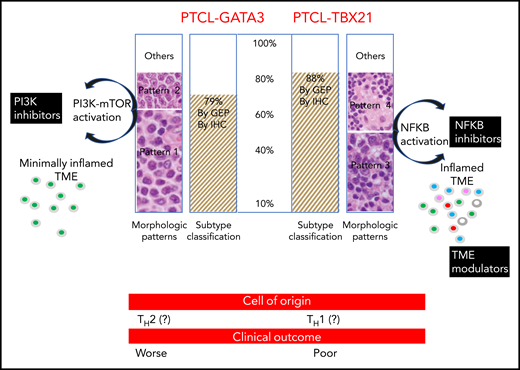
In this issue of Blood, Amador et al identified 2 distinct subtypes of peripheral T-cell lymphoma-not otherwise specified (PTCL-NOS) using gene expression profiling (GEP) and an immunohistochemistry (IHC) algorithm. The authors also showed that these lymphomas display distinct prognostic and morphologic features (see figure).1
PTCL-GATA3 and PTCL-TBX21 subtypes were found in ∼85% of cases defined by IHC and GEP. PTCL-GATA3 was associated with monomorphic patterns (histologic patterns 1 and 2), whereas PTCL-TBX21 was associated with polymorphic patterns (histologic patterns 3 and 4). Monomorphic patterns (patterns 1 and 2) were morphologically characterized by a monotonous tumor cell morphology with a minimal inflammatory TME. CD4+ T-cell rich, polymorphic patterns (patterns 3 and 4) were characterized by polymorphic neoplastic cells interspersed in a mixed inflammatory TME (pattern 3) or in the classic TME of lympho-histiocytic (Lennert) lymphoma (pattern 4). In the polymorphic patterns, the inflamed TME was CD4+ and CD8+ T-cell rich and contained eosinophils, plasma cells, and histiocytes. The PTCL-GATA3 subtype was associated with PI3K-mTOR activation, whereas PTCL-TBX21 showed NF-κB activation. Therefore, PTCL-GATA3 subtype patients may benefit from PI3K inhibitors, whereas PTCL-TBX21 subtype patients may benefit from NF-κB inhibitors and TME modulators. Significant differences in overall survival were observed between the PTCL-GATA3 and PTCL-TBX21 subtypes derived by IHC and GEP classification (not shown). CD4+ T cell CD8+ T cell eosinophil plasma cell histiocyte. The figure has been adapted from Figure 5 in the article by Amador et al that begins on page 2159.
PTCL-GATA3 and PTCL-TBX21 subtypes were found in ∼85% of cases defined by IHC and GEP. PTCL-GATA3 was associated with monomorphic patterns (histologic patterns 1 and 2), whereas PTCL-TBX21 was associated with polymorphic patterns (histologic patterns 3 and 4). Monomorphic patterns (patterns 1 and 2) were morphologically characterized by a monotonous tumor cell morphology with a minimal inflammatory TME. CD4+ T-cell rich, polymorphic patterns (patterns 3 and 4) were characterized by polymorphic neoplastic cells interspersed in a mixed inflammatory TME (pattern 3) or in the classic TME of lympho-histiocytic (Lennert) lymphoma (pattern 4). In the polymorphic patterns, the inflamed TME was CD4+ and CD8+ T-cell rich and contained eosinophils, plasma cells, and histiocytes. The PTCL-GATA3 subtype was associated with PI3K-mTOR activation, whereas PTCL-TBX21 showed NF-κB activation. Therefore, PTCL-GATA3 subtype patients may benefit from PI3K inhibitors, whereas PTCL-TBX21 subtype patients may benefit from NF-κB inhibitors and TME modulators. Significant differences in overall survival were observed between the PTCL-GATA3 and PTCL-TBX21 subtypes derived by IHC and GEP classification (not shown). CD4+ T cell CD8+ T cell eosinophil plasma cell histiocyte. The figure has been adapted from Figure 5 in the article by Amador et al that begins on page 2159.
Previous GEP studies have defined 2 major molecular subtypes within the group of PTCL-NOS diseases.2,3 One subtype was identified by the expression of GATA3 and its target genes and was designated as the PTCL-GATA3 subtype; the other was identified by the expression of T-box 21 (TBX21) and its target genes and was designated as the PTCL-TBX21 subtype.2,3 GATA3 is the transcriptional regulator in TH2 cell differentiation, whereas TBX21 is the regulator in TH1 and cytotoxic T-cell differentiation. Therefore, it has been hypothesized that PTCL-GATA3 and PTCL-TBX21 could originate from TH2 or TH1, respectively. Very recently, genetic studies have highlighted the role of distinct genetic pathways and enrichment of oncogenic pathways in the development of these lymphomas4 (see figure).
Starting from these GEP results, Amador et al have successfully generated an IHC algorithm with proven interobserver reproducibility and easy applicability to clinical practice for PTCL-NOS subclassification. Once the PTCL-NOS diagnosis has been made, 4 additional stains using commercially available antibodies for GATA3, CCR4, TBX21, and CXCR3 on formalin-fixed-paraffin-embedded tissue sections were recommended to recognize the 2 molecular subtypes. The PTCL-GATA3 and the PTCL-TBX21 subtypes identified by the IHC algorithm strongly matched those identified by the GEP results. Therefore, the IHC algorithm was suitable as a valid surrogate for GEP to subclassify PTCL-NOS. The study also showed that the PTCL-GATA3 and the PTCL-TBX21 subtypes exhibited distinct morphologic patterns and distinct tumor microenvironment (TME) compositions. However, the morphologic pattern was not integrated within the IHC algorithm. Furthermore, according to the conclusions of the study, the IHC algorithm could reliably facilitate risk stratification in future clinical trials, because these subtypes showed a difference in overall survival in affected patients.
Nowadays, approximately one third of PTCL cases cannot be classified further and are designated as PTCL-NOS.5 The research by Amador et al results in a classification proposal for this group of T-cell lymphomas that are currently undetermined.6 The major merit of this study is that the authors have translated the GEP signatures of PTCL-NOS into a clinically practical IHC algorithm that can identify 2 distinct subtypes of PTCL-NOS (ie, PTCL-GATA3 and PTCL-TBX21). The discovery of these subtypes improves the understanding of PTCL-NOS, patient management, and outcome. In fact, the PTCL subtypes that have been recognized by matching genomic and IHC approaches are also characterized by specific, yet different, pathogenetic pathways and TME composition (ie, inflamed vs minimally inflamed TME). Regardless of whether these are biological or pathological entities, what matters for the management of patients is that the oncogenic pathways found and the TME immunomodulators observed in the 2 PTCL subtypes are sensitive to targeted therapies7,8 (see figure). The choice of personalized treatment may also be guided by the analysis of TME to identify which cells or proteins are limiting the efficacy of therapy in individual patients. The immunomodulatory proteins expressed by the TME cell populations can be studied by using multiplex immunofluorescence or IHC assisted by digital image analysis.9 In addition, an in situ proximity ligation assay10 can be used for insight into possible protein-protein interactions.
In conclusion, the 2 molecular subsets of PTCL-NOS are distinct entities that should provide an advance in the classification of PTCL. It is evident that molecular profiling can refine existing classifications of T-cell lymphomas. Because these developments are having a profound impact on how lymphoma is diagnosed and treated, it is appropriate to define new lymphoma classes according to the mounting molecular genetic data.
Conflict-of-interest disclosure: The authors declare no competing financial interests

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal