Key Points
TET2 mutations confer a myeloid proliferation bias.
DNMT3A mutations occur in a multipotent stem cell.
Abstract
We analyzed DNA from polymorphonuclear (PMN) cells, monocytes, B cells, and T cells of 107 individuals with clonal hematopoiesis of indeterminate potential (CHIP) to perform lineage restriction analysis of different gene mutations. Three lineage categories were defined: myeloid (PMN with or without monocytes), myelolympho-B (myeloid and B cells), and multipotent (myeloid, B and T cells). Six individuals with aberrant patterns were excluded from analysis. Ninety-four had a single mutation (56 in DNMT3A, 24 in TET2, 7 in other genes [JAK2, ASXL1, CBL or TP53]). Fourteen had multiple mutations. The lineage restriction patterns of single DNMT3A- or TET2-mutated individuals were different. The proportion of myeloid restricted mutations was higher for TET2 (54.2%, 13 of 24) than for DNMT3A (23.2%, 13 of 56) (P < .05). It was similar for myelolympho-B category but with a 1.5 fold greater proportion of myeloid cells for TET2 individuals (P < .05). Importantly, 0% (0 of 24) of the individuals with TET2 mutation in the multipotent category in contrast to 35.7% (20 of 56) for DNMT3A (P < .01). The clone size predicted multipotent pattern for DNMT3A suggesting a time delay for extensive lineage clonal dominance. These distinctive features may be important in deciphering the transformation mechanisms of these frequent mutations.
Introduction
Age-related clonal hematopoiesis, which was initially suggested by X chromosome inactivation studies,1 is associated with acquired mutations in driver and nondriver genes.2-6 Age-related clonal hematopoiesis is highly prevalent after age 65 years and confers an increased risk of progression to hematologic cancers and cardiovascular complications.4-7 The precise risk associated with the presence of a mutation in a driver gene (eg, DNMT3A vs others) in healthy individuals is uncertain at this time. Thus, a provisional clinical entity named clonal hematopoiesis of indeterminate potential (CHIP) was created.8
Specific genes mutated in CHIP have been suggested to be founder events in the biology of myeloid cancers, such as myelodysplastic syndrome9 or acute myelogenous leukemia,10 and to be a potential reservoir for relapse.10 The identification of the cell of origin in which these mutations occur could have implications for diagnosis, prognosis, and even therapeutic strategies. Young et al11 looked at lineage restriction in a limited number of individuals, but no conclusive mutation-specific patterns could be identified, We recently documented that 93% of driver mutations in CHIP concerned DNMT3A or TET2, and they have demonstrated distinctive features such as greater age dependency, greater proliferation rate, and genetic predisposition for TET2.12 Herein we demonstrate different hematopoietic lineage involvement of mutations in these 2 genes in a cohort of 107 normal aging individuals with CHIP.
Study design
Cohort
We previously sequenced a cohort of 2530 normal aging individuals using the Ampliseq AML Panel (Thermo Fisher Scientific) covering 19 recurrently mutated genes in myeloid cancers12 and identified driver mutations in 347 individuals.12 Among these, 107 prospectively recruited individuals were selected on the basis of availability of DNA from polymorphonuclear (PMN) cells, monocytes, B cells, and T cells, thus allowing lineage restriction analysis (Figure 1A). All participants answered a medical questionnaire and gave informed consent. The study was approved by the Maisonneuve-Rosemont Hospital Ethics Committee in 1998 and was reapproved annually.
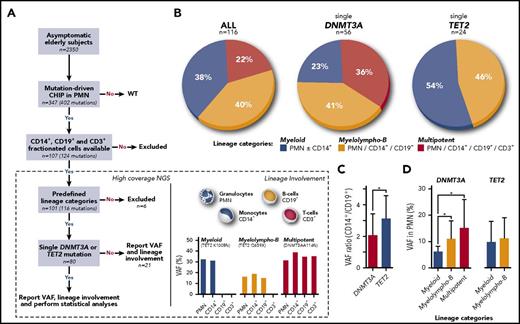
Lineage involvement. (A) Sample processing. (B) Distribution of lineage involvement patterns for all mutations (n = 116), single DNMT3A mutations (n = 56), and single TET2 mutations (n = 24). (C) CD14+:CD19+ VAF ratio in DNMT3A (n = 23) and TET2 (n = 11) individuals with myelolympho-B mutation. (D) Mean VAF for myeloid, myelolympho-B, and multipotent DNMT3A and TET2 mutations. *P < .05. NGS, next-generation sequencing; WT, wild-type.
Lineage involvement. (A) Sample processing. (B) Distribution of lineage involvement patterns for all mutations (n = 116), single DNMT3A mutations (n = 56), and single TET2 mutations (n = 24). (C) CD14+:CD19+ VAF ratio in DNMT3A (n = 23) and TET2 (n = 11) individuals with myelolympho-B mutation. (D) Mean VAF for myeloid, myelolympho-B, and multipotent DNMT3A and TET2 mutations. *P < .05. NGS, next-generation sequencing; WT, wild-type.
Sample processing
Blood cells were obtained by venipuncture. Complete blood counts were obtained from a GenS automated cell counter (Beckman Coulter). Blood cells were separated into PMN and mononuclear fractions by standard density gradient centrifugation (Ficoll-Paque). Fluorescence-activated cell sorting was used to isolate monocytes (CD14+ cells), B cells (CD19+ cells), and T cells (CD3+ cells) from the mononuclear cell fraction (FACStar Plus, Becton Dickinson, San Jose, CA).
Lineage restriction categories
The 3 lineage restriction categories are myeloid (mutations detected in PMN with or without monocytes), myelolympho-B (mutations detected in myeloid and B cells), and multipotent (mutations detected in myeloid, B, and T cells) (Figure 1A).
Sequencing and analysis
In all, 428 DNA samples (PMN, CD14+, CD19+, and CD3+ cell fraction) from 107 individuals were sequenced at high coverage (95% >500×) on an Ion Proton sequencer using the Ampliseq AML Panel (Thermo Fisher Scientific) as described in Buscarlet et al.12 All mutations were visually inspected on an Integrative Genomics Viewer. Samples were considered mutated if the mutation was detected above a variant allele frequency (VAF) of 2%.
Statistical analyses
The descriptive statistics and analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC). For binary outcomes, generalized estimating equations were used to adjust for age and family structure except when comparing multipotent with non-multipotent groups, in which a Fisher’s exact test without covariates was used because of low counts. Quantitative traits were log transformed and analyzed with linear mixed models adjusting for age and family as random effects. Blood counts were adjusted for age and smoking.
Results and discussion
The study cohort comprised individuals age 55 to 96 years (mean age, 70 years). Sixty-two individuals had a single DNMT3A mutation (mean VAF in PMN, 11.2%), 24 had a single TET2 mutation (mean VAF in PMN, 10.5%), 7 had a single mutation in other genes (JAK2, ASXL1, CBL, or TP53), and 14 had multiple mutations. Individuals with multiple mutations were older than those with a single mutation (74.8 vs 69.2 years; P = .05), and individuals with TET2 mutations were older than those with DNMT3A mutations (72.3 vs 67.7 years; P = .04). Sequence analyses in all cells types were available for the 107 participants. A small number of participants had phenotypes that did not correspond to the predefined lineage categories, so they were removed from analysis. These aberrant patterns were largely a result of mutations with a VAF lower than 2% in expected lineages. The distribution of the mutations in the remaining 101 participants, according to the predefined lineage categories, was myeloid, 44; myelolympho-B, 47; and multipotent, 25 (Figure 1B).
We then compared lineage involvement between individuals with single DNMT3A or TET2 mutations. Important differences were noted. First, the proportion of myeloid-restricted mutations was higher for individuals with TET2 (54.2%; 13 of 24) than for those with DNMT3A (23.2%; 13 of 56) (P = .01). Second, the proportion of individuals with myelolympho-B was relatively similar (45.8% vs 41.1%) between the 2 groups, but the CD14+:CD19+ VAF ratio was higher for TET2 (2.96; n = 11) than for DNMT3A (1.73; n = 23; P = .05; Figure 1C), suggesting that TET2 mutations exert a stronger myeloid bias than DNMT3A mutations. More importantly, there was not a single TET2-mutated individual who had this mutation in CD3+ cells. In contrast, 35.7% (20 of 56) of DNMT3A-mutated individuals met the criteria for multipotent involvement (P < .01; Figure 1B). The lineage restriction of DNMT3A individuals was not influenced by the type of mutation (data not shown). This is in line with a mouse model of dnmt3a.13 The absence of evidence of multipotent involvement for TET2 was surprising because TET2 has been involved in lymphoma predominantly of the T-cell type.14 The simplest explanation is that TET2 mutations can occur independently in myeloid- or lymphoid-restricted hematopoietic stem cells (HSCs) in different individuals (Figure 2). Alternatively, TET2 mutations could occur directly in a multipotent HSC, but cooperation with secondary mutations or epigenetic modifications may influence lineage proliferation bias. If this is the case, these secondary events would be different between TET2 and DNMT3A. The data obtained for the 7 participants with mutations in genes other than DNMT3A or TET2 were heterogeneous but none had the multipotent pattern. One important observation is that for both DNMT3A and TET2, we were able to demonstrate a close relationship between myeloid and B-cell proliferation. This is in agreement with the recent description of clonal output of transplanted barcoded HSCs of rhesus macaques15 and the revisited hematopoietic hierarchy models.16-18
Model of DNMT3A and TET2 clonal origin in CHIP.DNMT3A and TET2 mutations have different lineage involvement. This is compatible with DNMT3A mutations occurring in a multipotent HSC and TET2 mutations in a more committed HSC. Alternatively, DNMT3A and TET2 mutations may occur in a multipotent HSC, but lineage-specific proliferation is influenced by different intrinsic/extrinsic factors conferring a strong myeloid bias for TET2-mutated HSCs.
Model of DNMT3A and TET2 clonal origin in CHIP.DNMT3A and TET2 mutations have different lineage involvement. This is compatible with DNMT3A mutations occurring in a multipotent HSC and TET2 mutations in a more committed HSC. Alternatively, DNMT3A and TET2 mutations may occur in a multipotent HSC, but lineage-specific proliferation is influenced by different intrinsic/extrinsic factors conferring a strong myeloid bias for TET2-mutated HSCs.
We then questioned whether the time of evolution of the mutated clone could have influenced the results. For example, if a mutation occurs in a multipotent HSC, the amount of time it takes to affect myeloid, B, and T cells might be different, T cells being the longest lived of the 3 cell types. Although our investigation is limited to a single sampling point, we hypothesized that there is a temporal and proportional relationship between time and clone size measured by the VAF. For DNMT3A, we demonstrated that VAF increases proportionally to the extent of lineage involvement categories: myeloid mean VAF, 5.8%; myelolympho-B mean VAF, 9.5%; and multipotent category mean VAF, 12.0% (P < .05; Figure 1D). Likewise, selecting participants with a VAF >10% in PMN cells almost completely eliminated myeloid restriction for DNMT3A mutations. This suggests that DNMT3A mutation occurs in a multipotent HSC in the majority of participants but that lymphoid lineage involvement is time dependent and/or influenced by other variables acting on the kinetics of clonal expansion. No such correlation was seen with TET2 in which VAF in myeloid or myelolympho-B categories were similar. We previously found that DNMT3A and TET2 mutations in PMN did not have an impact on blood counts compared with those for individuals of comparable age who had no mutations.12 In this study, we documented that specific lineage categories have no impact on blood parameters.
In conclusion, we demonstrated that DNMT3A and TET2, the 2 most frequently mutated genes in CHIP, have distinctive patterns of hematopoietic cell involvement and a different myeloid proliferation bias. Similar results have recently been reported by Arends et al.19 This adds to other features that we have previously identified such as greater influence of age and greater proliferation rate and heritability for TET2.12 These differences support that CHIP is a broad clinical entity and that gene-specific approaches are needed to understand and decipher the mechanisms associated with progression to hematologic cancers or other medical conditions.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research and by the Leukemia Lymphoma Society of Canada.
Authorship
Contribution: M.B. performed and analyzed all experiments, interpreted data, planned the statistical analyses, generated all figures, and cowrote the manuscript; V.B. provided technical support; M.-P.D. supervised the statistical analyses; S.P. performed the statistical analyses with Y.F.Z.; L.M. revised the manuscript; and L.B. conceived the study, interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lambert Busque, Hôpital Maisonneuve-Rosemont, Montréal, QC H1T 2M4, Canada; e-mail: lbusque.hmr@ssss.gouv.qc.ca.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal