To the editor:
Multiple myeloma (MM) is characterized by a large diversity of genetic abnormalities.1-3 They can be classified in 3 categories: copy number changes, mutations, and translocations involving mainly the IGH gene at 14q32. Translocations are usually balanced translocations with various partner genes: CCND1 at 11q13 (15%-20% of the patients), MMSET/FGFR3 at 4p16 (12%-15%), MYC at 8q24 (15%), MAF at 16q23 (3%), MAFB at 20q11 (1%), and CCND3 at 6p21 (1%). Several papers characterized the t(4;14) and t(14;16), but few analyzed the most frequent 1, t(11;14),4,5 mainly because, in contrast to the other 2, which are associated with a poor prognosis,6 the t(11;14) was not associated with a specific outcome until recently. However, recent reports suggest that outcome of patients displaying t(11;14) is inferior to other standard risk patients.7,8 The development of novel strategies and drugs in the past decades dramatically improved the survival of MM patients.9-12 The next step would be to define the right treatment of the right patient. In line with this goal, translocation t(11;14) is becoming more interesting for physicians with the demonstration of a specific good response (40% of overall response rate) with venetoclax, a BCL-2 inhibitor.13,14 Even if direct comparison cannot be done because recent clinical trials did not report specific t(11;14) response rate, these results obtained in heavily pretreated relapsed/refractory with an oral single agent are better than expected.
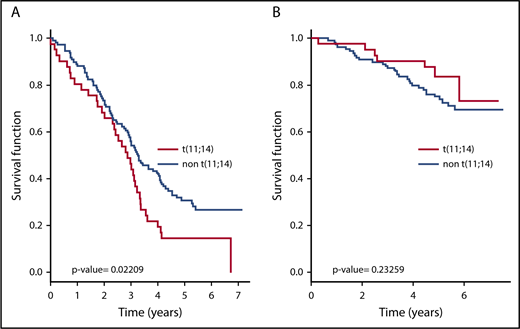
We thus decided to characterize the transcriptomic profile of a large number of patients with t(11;14). All patients provided signed consent for these genetic analyses in accordance with the Declaration of Helsinki. This study was approved by Toulouse Ethic Committee. RNA sequencing experiments (with tumor purity >90%) were conducted on 157 patients at diagnosis within the Randomized Study Comparing Conventional Dose Treatment Using a Combination of Lenalidomide, Bortezomib and Dexamethasone to High-Dose Treatment With ASCT in the Initial Management of Myeloma in Patients up to 65 Years of Age 2009 trial for which fluorescence in situ hybridization was performed, allowing the identification of 43 (27%) patients with a t(11;14). All patients received pretransplant bortezomib-lenalidomide-dexamethasone induction. In accordance to previous reports4,5 supporting a standard risk profile of t(11;14), a 5-year median follow-up of our patients could not associate the presence of this translocation with a specific outcome (Figure 1). By contrast, a recent study8 reported a subgroup t(11;14) displaying a significantly shorter survival than a non-14q32 translocation group. The very nature of this comparison, added to the demographical and treatment features (only 60% were transplant eligible, and most patients received 1 novel agent-based induction [proteasome inhibitor or Immunomodulatory imide drugs]), may explain this different interpretation. Of note, in our study, the incidence of 17p deletion, 1q gain, and 1p32 deletion were comparable between t(11;14) and non-t(11;14) subgroups. We then identified a specific transcriptomic signature with 2345 differentially expressed genes in patients displaying t(11;14) compared with the rest of the patients. As expected, the most discriminant gene was CCND1 (Figure 2A), which was highly expressed in patients with t(11;14). However, 11 genes from the apoptosis family were differentially expressed. These included 2 members of the BCL2 family, BCL2L1 and BAK1, that were underexpressed by t(11;14) patients (Figure 2B), 7 members of the tumor necrosis factor (TNF) receptor superfamily (TNFRSF18, TNFRSF4, FAS, TNFRSF10B, TNFRSF10D, and TNFRSF13C were overexpressed, and TNFRSF17 was underexpressed) and 2 members of the BH3-only family, namely PMAIP1 [NOXA] which was overexpressed, and BIK which was underexpressed in the patients with t(11;14). Interestingly, no member of the caspase family or other apoptotic genes was differentially expressed.
Survival of t(11;14) myeloma patients. Analysis was done with 40 t(11;14) patients and 110 non-t(11;14) patients. The median follow-up was 5 years. (A) Event-free survival. (B) Overall survival.
Survival of t(11;14) myeloma patients. Analysis was done with 40 t(11;14) patients and 110 non-t(11;14) patients. The median follow-up was 5 years. (A) Event-free survival. (B) Overall survival.
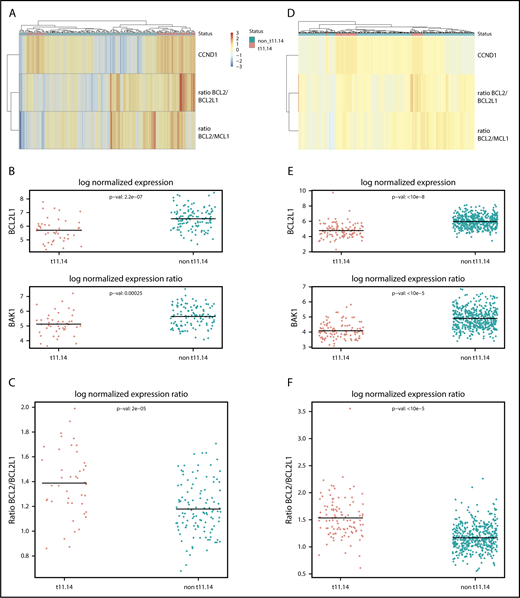
Transcriptomic comparison of t(11:14) and other MM patients. (A-C) The Randomized Study Comparing Conventional Dose Treatment Using a Combination of Lenalidomide, Bortezomib and Dexamethasone to High-Dose Treatment With ASCT in the Initial Management of Myeloma in Patients up to 65 Years of Age 2009 cohort and (D-F) the independent Multiple Myeloma Research Foundation cohort. (A,D) Heat map of the normalized expression of CCND1 and BCL2/BCL2L1 and BCL2/MCL1 expression ratio in the cohorts. The t(11;14) patients (indicated in pink on the first line) cluster in 2 distinct groups based on their BCL2/BCL2L1 ratio. (B,E) Normalized expression of differentially expressed genes from the apoptosis family between t(11;14) and non-t(11;14) patients. (C,F) Normalized expression ratio of BCL2/BCL2L1 between t(11;14) and non-t(11;14) patients.
Transcriptomic comparison of t(11:14) and other MM patients. (A-C) The Randomized Study Comparing Conventional Dose Treatment Using a Combination of Lenalidomide, Bortezomib and Dexamethasone to High-Dose Treatment With ASCT in the Initial Management of Myeloma in Patients up to 65 Years of Age 2009 cohort and (D-F) the independent Multiple Myeloma Research Foundation cohort. (A,D) Heat map of the normalized expression of CCND1 and BCL2/BCL2L1 and BCL2/MCL1 expression ratio in the cohorts. The t(11;14) patients (indicated in pink on the first line) cluster in 2 distinct groups based on their BCL2/BCL2L1 ratio. (B,E) Normalized expression of differentially expressed genes from the apoptosis family between t(11;14) and non-t(11;14) patients. (C,F) Normalized expression ratio of BCL2/BCL2L1 between t(11;14) and non-t(11;14) patients.
Based on the recent encouraging clinical activity of the BCL-2 inhibitor venetoclax in t(11;14) MM, with a good correlation with high BCL2/BCL2L1 and/or BCL2/MCL1 messenger RNA ratio, we analyzed these ratios in this population. Although the BCL2/MCL1 did not discriminate the 2 populations, overall the BCL2/BCL2L1 ratio was generally significantly higher in t(11;14) patients (Student 2-sided P = 2 × 10−5; Figure 2A,C). The BCL2/BCL2L1 ratio separates the t(11;14) patients in 2 groups, with only two-thirds of the patients (30/43) displaying a high ratio despite all patients having high CCND1 expression. This separation does not correlate with the CD-1/CD-2 molecular classification of the CCND1/t(11;14) myeloma patients,15,16 and BCL2L1 was the only gene whose expression differentiates both groups. Interestingly, high BCL2/BCL2L1 ratio was frequently identified in a subgroup of non-t(11;14) patients as well (Figure 2A), with a transcriptomic signature of >3000 genes characterizing those high-ratio patients. Although BCL2L1 and BCL2 were the most differentially expressed genes, it is likely that this signature is a result of a large heterogeneity of patients in the groups. These results were confirmed by the independent study of the Multiple Myeloma Research Foundation RNA-sequencing cohort (performed on 564 patients, including 104 patients with t(11;14); , the BCL2/BCL2L1 ratio in this cohort being significantly higher [P = 2 × 10−16]; Figure 2D-F). BCL2L1 encodes the anti-apoptotic Bcl-XL protein, whose expression has been correlated to resistance to venetoclax in a preclinical MM model.17
So, which marker best predicts response to venetoclax? Because our patients were not treated with venetoclax, we cannot answer this important question. Our data may explain the better responses in t(11;14) patients, but not in all of them. In contrast, some patients lacking the t(11;14) also display high ratios and low BCL2L1 or BAK expression. These patients might be good candidates for venetoclax treatment. We believe that these issues should be addressed specifically in the ongoing and upcoming venetoclax trials by comparing transcriptome from plasma cells obtained in responder vs nonresponder patients. If 1 predictive marker is actually identified, a more accessible method will have to be developed for clinical use, such as flow cytometry or reverse transcription polymerase chain reaction, making the opportunity to practice targeted-based precision medicine in multiple myeloma.
In summary, we confirm that t(11;14) is not associated with a specific outcome, but this may change if clinical efficacy of venetoclax is confirmed in this subgroup. A high ratio BCL2/BCL2L1 or low expression of BCL2L1 or BAK might select patients with a high probability of response. Further studies are needed to establish a potential link with response to venetoclax.
Acknowledgments
The authors thank the Intergroupe Francophone du Myélome for providing patient samples and clinical data.
This work was supported by National Institutes of Health, National Cancer Institute grants PO1-155258 and P50-100707 (H.A.L.) and the Cancer Pharmacology of Toulouse and Region (CAPTOR) program. The Centre de Recherches en Cancérologie de Toulouse Team 13 is supported by la Fondation ARC (Association pour la Recherche sur la Cancer, grant PGA1*20160203788).
Authorship
Contribution: A.C., N.M., H.A.-L., and J.C. designed research, analyzed data, and wrote the manuscript, which was reviewed and edited by the other coauthors; L.B. and S.M. performed laboratory work; A.C. and M.S. analyzed the data; and A.P., M.F., and M.A. provided samples and clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jill Corre, Unit for Genomics in Myeloma, Institut Universitaire du Cancer-Oncopole, 1 Avenue Irène Joliot-Curie, 31100 Toulouse, France; e-mail: corre.jill@iuct-oncopole.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal