Abstract
Our knowledge of myeloma genetics remained limited and lagged behind many other hematologic malignancies because of the inherent difficulties in generating metaphases within the malignant plasma cell clone. With the development of molecular techniques (microarrays and next-generation sequencing), our understanding has been highly improved in the past 5 years. These studies have not only confirmed the prevalence of wide heterogeneity in myeloma at the molecular level, but has also provided a much clearer picture of the disease pathogenesis and progression. Whether these data will enable improvements in the therapeutic approach is still a matter of debate. The next improvement will come from detailed analyses of these molecular features to try to move from a treatment fitted to every patient to individualized therapies, taking into account the complexity of the chromosomal changes, the mutation spectrum, and subclonality evolution.
Introduction
Multiple myeloma (MM) is a heterogeneous hematologic malignancy that occurs mainly in the elderly population (median age at diagnosis ∼70 years). Because of major improvements in the general care of patients over the past 50 years, leading to a marked increase in longevity, the incidence of MM is increasing worldwide. It is currently accepted that all MM cases are preceded by an asymptomatic expansion of clonal plasma cells, known as monoclonal gammopathy of undetermined significance (MGUS), and smoldering MM (SMM).1,2 A fraction of these individuals with MGUS or SMM will evolve to symptomatic MM, but most of the MGUS cases will remain totally asymptomatic. Symptomatic MM is clinically characterized by lytic bone disease, anemia, hypercalcemia, renal failure, and susceptibility to bacterial infections. Why some MGUSs will remain totally asymptomatic for decades whereas others will evolve to overt MM is currently unknown, but the main hypothesis is the occurrence of “malignant” genetic events in evolving patients. To understand these events, a large amount of work has been dedicated to dissect the oncogenesis of MM.
Cell of origin
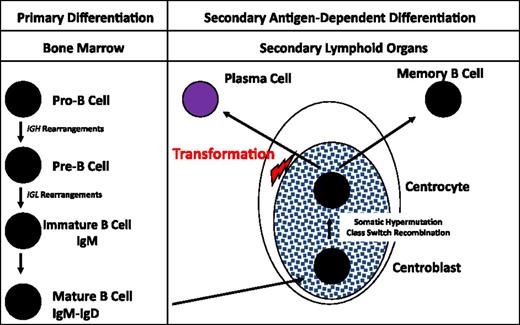
Plasma cells represent the final differentiation stage of B cells. The first steps of differentiation occur within the bone marrow. At the molecular level, the first steps of this differentiation process are the rearrangements of the heavy chain immunoglobulin (Ig) gene (IGH). This gene is a very large gene (∼2 Mb), presenting 4 major domains: the variability domain (VH, containing more than 100 DNA segments), the diversity domain (DH, containing 27 DNA segments), the joining domain (JH, containing 6 DNA segments), and the constant domain (containing 9 DNA segments). The first rearrangements are DNA deletions combining 1 DH segment to 1 of the 6 JH segments. These deletions are supposed to be stochastic, independently of any antigen pressure. If molecularly productive, the pro-B cell continues its differentiation by combining this DH-JH segment with a VH segment. These rearrangements are made and regulated by a specific recombinase enzyme, the recombination activating genes (RAG), which recognizes specific DNA motifs within the DH, JH, and VH segments. If these rearrangements are in frame, or “productive,” the pre-B cell will then rearrange the light chain genes, IGLκ and IGLλ. It first attempts to rearrange the IGLκ gene. If productive, the mature B cell will then be able to produce IgMκ, which is expressed at the B-cell surface. If unsuccessful (mainly by non–in-frame rearrangements), the B cell will then rearrange the IGLλ gene, leading to the production of an IgMλ. This process explains the disequilibrium in the type of B cells, two-thirds expressing an IgMκ at the membrane. These mature B cell will then quit the bone marrow to colonize the secondary lymphoid organs to continue its maturation. This second part of differentiation will become antigen-dependent, in relationship with dendritic and T cells. Within the germinal centers of the secondary lymphoid organs, a second type of molecular rearrangement will occur, known as the somatic hypermutation (SMH) process. Stochastic mutations will be produced within the VDJ segment by a specific enzyme, activation-induced deaminase. Only B cells with mutations improving the specificity of the antibody for the antigen will survive, the others dying via apoptosis. The last rearrangement process also occurs in the secondary lymphoid organs and is known as the class switch recombination (CSR). During this process, specific DNA segments known as switch regions will be recombined on the dependence of the activation-induced deaminase enzyme, with deletion of the interswitch region DNA. The mature B cell will then express a different Ig, either IgG, IgA, or IgE. Finally, these mature B cells will either differentiate in memory B cells or in long-lived plasma cells, which will return to bone marrow.
The oncogenic transformation in MM is thought to occur within these secondary lymphoid organs. Several pieces of evidence support this hypothesis (Figure 1). First, malignant plasma cells present a high rate of somatic mutations, with no heterogeneity, suggesting that the oncogenetic event occurred after the end of the SMH process, which physiologically takes place in the germinal centers of secondary lymphoid organs. The second piece of evidence is the nature of the monoclonal Ig, essentially IgG and IgA, rarely IgD or IgM. Here again, the CSR process is supposed to occur in the germinal centers. Finally, the molecular analyses of some of the oncogenic events, and especially of recurrent chromosomal translocations involving the IGH gene (see “Oncogenesis”), showed that the t(4;14) largely involves the switch regions, suggesting errors during the CSR process,3,4 whereas the t(11;14) may result from errors during the SMH one.4 A recent paper from the UK group suggested that some of the IGH translocations may take place within the bone marrow during the maturation IGH rearrangements.4 This hypothesis is somewhat controversial taking into account all the pieces of evidence of a postgerminative tumor. One alternative hypothesis could be the occurrence of RAG-mediated molecular errors during a (re-)edition of the B-cell receptor. This second hypothesis (difficult to demonstrate) would have the advantage of a unifying model of oncogenesis. A recent study utilizing deep IgH sequencing in myeloma cells demonstrated >4% of patients having 2 unrelated clones with different VDJ rearrangements. This raises an intriguing possibility of evolution of 2 independent clones at an earlier stage of plasma cells before VDJ rearrangement. Additionally, a small proportion of patients had related IgH clones suggesting continued SMH (Table 1).5
B-cell differentiation. The first step of B-cell engagement is characterized by an IGHDH-JH rearrangement, followed by a VH-DH-JH fusion. If “productive” (or successful), these IGH rearrangements are followed by recombinations of the IGLκ, and/or IGLλ genes. These DNA rearrangements take place within the bone marrow and are totally antigen independent. The B cells then migrate to the secondary lymphoid organs where, within the germinal centers, they terminate their differentiation through the SMH and CSR processes. This differentiation step is antigen dependent, in cooperation with dendritic and T cells. The oncogenetic event is supposed to take place after this long differentiation process, before the migration of the plasma cell to bone marrow.
B-cell differentiation. The first step of B-cell engagement is characterized by an IGHDH-JH rearrangement, followed by a VH-DH-JH fusion. If “productive” (or successful), these IGH rearrangements are followed by recombinations of the IGLκ, and/or IGLλ genes. These DNA rearrangements take place within the bone marrow and are totally antigen independent. The B cells then migrate to the secondary lymphoid organs where, within the germinal centers, they terminate their differentiation through the SMH and CSR processes. This differentiation step is antigen dependent, in cooperation with dendritic and T cells. The oncogenetic event is supposed to take place after this long differentiation process, before the migration of the plasma cell to bone marrow.
Controversial issues in genetics of MM
| Assumption . | Controversy . | Hypothesis/question/exploration . |
|---|---|---|
| The t(4;14) largely involves the switch regions, suggesting errors during the CSR process. | Some of the IGH translocations may take place not in secondary lymphoid organs but within the bone marrow during the maturation IGH rearrangements | Occurrence of RAG-mediated molecular errors during a (re-)edition of the B-cell receptor. |
| The oncogenic transformation in MM is thought to occur within secondary lymphoid organs. | More than 4% of patients have 2 unrelated clones with different VDJ rearrangements. | Possibility of evolution of 2 independent clones at an earlier stage of plasma cells before VDJ rearrangement. |
| Hyperdiploidy is probably because of missegregation of chromosomes during mitosis. | The nature of the gained chromosomes in hyperdiploidy is totally different between ALL and MM. | Do chromosomes have a specific disposition on the mitotic plaque in MM as compared with ALL, leading to gains of different chromosomes? |
| Some of the 14q32 translocations are recurrent. | The selectivity in the nature of 14q32 translocation partners is not fully understood. | The vicinity of chromosomal domains of chromosomes 14, 11, and 4 at certain times of SMH or CSR. |
| Several subclones coexisting at the same time in a single patient are similarly distributed in all locations. | Several subclones coexisting at the same time in a single patient have differential locations. | Several aspirates/biopsies should be performed in a single patient, ideally MRI- or PET-CT-guided on focal lesions, with subsequent evaluation for clonal content and subclone distribution |
| The subclone selection is under therapeutic pressure. | The subclone selection occurs in the natural history of the disease, which includes growth potential of the subclones or their interaction with their microenvironment. | Data supporting both hypotheses. The only way to answer this question would be to systematically analyze the diagnostic and first relapse specimens in a homogeneously treated cohort of patients. |
| Rare recurrent mutations observed only in subclones include genes supposed to act as drivers, such as NRAS, KRAS, and BRAF. | Driver mutations are not always expressed at the RNA level. | Need for future analyses at the protein level and to define the role of some of the known DNA repair mechanisms in inducing clonal change. |
| Assumption . | Controversy . | Hypothesis/question/exploration . |
|---|---|---|
| The t(4;14) largely involves the switch regions, suggesting errors during the CSR process. | Some of the IGH translocations may take place not in secondary lymphoid organs but within the bone marrow during the maturation IGH rearrangements | Occurrence of RAG-mediated molecular errors during a (re-)edition of the B-cell receptor. |
| The oncogenic transformation in MM is thought to occur within secondary lymphoid organs. | More than 4% of patients have 2 unrelated clones with different VDJ rearrangements. | Possibility of evolution of 2 independent clones at an earlier stage of plasma cells before VDJ rearrangement. |
| Hyperdiploidy is probably because of missegregation of chromosomes during mitosis. | The nature of the gained chromosomes in hyperdiploidy is totally different between ALL and MM. | Do chromosomes have a specific disposition on the mitotic plaque in MM as compared with ALL, leading to gains of different chromosomes? |
| Some of the 14q32 translocations are recurrent. | The selectivity in the nature of 14q32 translocation partners is not fully understood. | The vicinity of chromosomal domains of chromosomes 14, 11, and 4 at certain times of SMH or CSR. |
| Several subclones coexisting at the same time in a single patient are similarly distributed in all locations. | Several subclones coexisting at the same time in a single patient have differential locations. | Several aspirates/biopsies should be performed in a single patient, ideally MRI- or PET-CT-guided on focal lesions, with subsequent evaluation for clonal content and subclone distribution |
| The subclone selection is under therapeutic pressure. | The subclone selection occurs in the natural history of the disease, which includes growth potential of the subclones or their interaction with their microenvironment. | Data supporting both hypotheses. The only way to answer this question would be to systematically analyze the diagnostic and first relapse specimens in a homogeneously treated cohort of patients. |
| Rare recurrent mutations observed only in subclones include genes supposed to act as drivers, such as NRAS, KRAS, and BRAF. | Driver mutations are not always expressed at the RNA level. | Need for future analyses at the protein level and to define the role of some of the known DNA repair mechanisms in inducing clonal change. |
ALL, acute lymphoblastic leukemia; CT, computed tomography; MRI, magnetic resonance imaging; PET, positron emission tomography.
Oncogenesis
The current widely accepted model of oncogenesis describes 2 different pathways.6 The first one is represented by hyperdiploidy, which is observed in up to 55% of the patients.7-11 Of note, the gained chromosomes are not random but involve specifically chromosomes 3, 5, 7, 9, 11, 15, 19, and 21. When compared with another B-cell malignancy (ie, childhood ALL), which also presents with hyperdiploidy, the nature of the gained chromosomes is totally different.12 How can this striking difference in 2 B-cell malignancies, 1 immature (ALL) and the other mature (MM), be explained? Neither experimental demonstration nor a hypothesis has been proposed so far to explain the pathogenetic role of this observation. Hyperdiploidy is probably because of missegregation of chromosomes during mitosis. Do chromosomes have a specific disposition on the mitotic plaque in MM as compared with ALL, leading to gains of different chromosomes? No answer can be proposed today. The second pathway is based on IGH translocations, observed in ∼40% to 50% of the patients and almost all the human myeloma cell lines (HMCLs).13-15 Some of these 14q32 translocations are recurrent, others being apparently random.16 The mechanisms have been described before, and the partner genes under dependence of the strong IGH enhancers, leading to overexpression of the targeted proteins. The most frequent one is the t(11;14)(q13;q32), observed in 15% to 20% of the patients and in 25% of the HMCLs.17 The t(11;14) dysregulates the CCND1 gene, leading its overexpression. In frequency, the second recurrent IGH translocation is the t(4;14)(p16;q32), observed in 12% to 15% of the patients and 25% of the HMCLs. This translocation is very peculiar in B-cell malignancies, by disrupting and upregulating 2 genes, FGFR3 and MMSET/WHSC1, creating a fusion transcript with this latter gene.18,19 FGFR3 is an oncogene activated by mutations in several solid tumor types. Of note, FGFR3 is upregulated only in 70% of the patients with the t(4;14), because of an unbalanced translocation with loss of the telomeric part of chromosome 4, bearing FGFR3.20-22 This finding suggests that the main molecular target of the translocation is MMSET. MMSET is a methyl-transferase protein, and its upregulation leads to methylation of several proteins in the genome.23 Other recurrent translocations are much more rarely observed, in <3% of the patients, such as the t(14;16)(q32;q23), which dysregulates the MAF oncogene24,25 ; the t(14;20)(q32;q11), dysregulating the MAFB oncogene26 ; or the t(6;14)(p21;q32), which upregulates the CCND3 gene.27 Of note, the t(14;16), if rare in patients, is present in 25% of the HMCLs, possibly because of its propensity to present as a plasma cell leukemia in patients. This selectivity in the nature of 14q32 translocation partners is not fully understood. The major hypothesis is the vicinity of domains of chromosomes 14, 11, and 4 at certain times of SMH or CSR.
Besides these supposedly primary events, many other chromosomal rearrangements are present in the tumor plasma cells at the time of diagnosis. The most frequent ones are monosomy 13 (45% of the patients),28 duplication of the long arm of chromosome 1 (1q gains, 30% to 35% of the patients),29 and different deletions involving the 1p, 6q, 8p, 12p, 14q, 16q, 17p, or 20p chromosomal regions.30 All these abnormalities have been known for a long time because they are visible on the conventional karyotype. More recent data based on the comparative genomic hybridization (CGH) or single nucleotide polymorphism (SNP)–array technologies have revealed other important chromosomal changes, especially homozygotic deletions. Several of these double deletions target inhibitors of the nuclear factor κB pathway, such as BIRC2/3 on chromosome 11, TRAF3 on chromosome 14, or CYLD on chromosome 16.31,32 Another recurrent double deletion is observed in a few percent of the patients on chromosome 1, targeting the tumor suppressor CDKN2C gene.33 Molecular analyses based on fluorescence in situ hybridization (FISH) or CGH/SNP-array revealed frequent chromosomal changes (translocations, deletions, duplications, insertions, and amplifications) in the 8q24 region.34 The molecular target of these rearrangements is the MYC oncogene, which is a key factor of MM biology. These data may be the chromosomal substratum of the frequent MYC upregulation observed at the RNA level in patients.
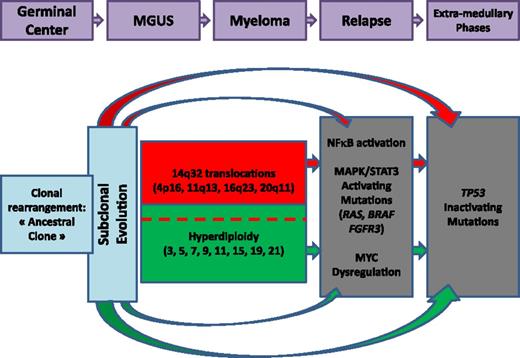
All these latter chromosomal changes are supposed to be secondary events in the MM oncogenesis. The main reason supporting this hypothesis is that most of them are often observed only in subclones, in contrast to IGH translocations, which are usually present in virtually 100% of the plasma cells. Taken together, all these genomic data have begun to enable us to build an oncogenetic model. Moreover, an important aspect that we need to consider is the observed genomic changes in MGUS/SMM and those acquired later with progression to active myeloma. This model, proposed several years ago, has been recently enriched by the subclonality concept (Figure 2; see “The subclonality concept”).
Oncogenetic model. Quickly after the oncogenetic event(s), differential mutations occur, creating subclones. The 2 main oncogenetic pathways are the “trisomy” pathway and the 14q32 translocation pathway. Secondary events occur later during evolution.
Oncogenetic model. Quickly after the oncogenetic event(s), differential mutations occur, creating subclones. The 2 main oncogenetic pathways are the “trisomy” pathway and the 14q32 translocation pathway. Secondary events occur later during evolution.
The subclonality concept
MM is a clonal malignancy characterized by the secretion of a monoclonal Ig or monoclonal free light chains. This dogma is (and will remain) accurate. However, detailed analyses at the molecular level have revealed a more complex situation. In 2012, 3 publications reported a certain degree of heterogeneity within the tumor clone, by comparing DNA abnormalities present at diagnosis and at relapse.35-37 Based on SNP array or high-throughput sequencing, all 3 reports demonstrated that the clone observed at relapse could be slightly different from the one observed at diagnosis. In some cases, the relapse clone presented more chromosomal/genetic changes, but on the same background, representing a classical clonal evolution. More strikingly, in some patients, the relapse clone presented pieces of DNA, which were absent at the time of diagnosis. Because ex nihilo creation of DNA is impossible, the conclusion of these studies was that minor subclones, undetectable at diagnosis by classical methods, were responsible for relapse. The DNA abnormalities defining these subclones were mainly supposed to be secondary changes. However, a study focused on the t(4;14), supposed to be a primary driver genetic event, showed that this translocation could be present only in a tiny minor subclone at diagnosis, but be the major clone at relapse, and vice versa.38 Thus, the development of subclones is a very early event in oncogenesis, probably soon after the cell transformation (Figure 3). An unresolved question is whether several subclones coexisting at the same time in a single patient have differential locations. This situation is well described in solid tumors, but no data are currently available in MM. To address this issue, several aspirates or biopsies should be performed in a single patient, ideally MRI- or PET-CT-guided on focal lesions, with subsequent high-throughput sequencing on all individual samples. Another unresolved question is mechanism of selection of subclones. Does the selection occur solely under therapeutic pressure, or does it occur in the natural history of the disease, which includes growth potential of the subclones or their interaction with their microenvironment? Actually, data supporting both hypotheses exist. A first study based on SNP array found that the large majority of “branching” evolutions (occurrence of a different subclone at the time of relapse) occur in patients treated by a bortezomib-dexamethasone combination as compared with a classical chemotherapeutic combination (vincristine-adriamycin-dexamethasone), suggesting that vincristine-adriamycin-dexamethasone displayed a broader activity on all subclones.35 The second example came from a single patient treated both frontline and at first relapse by a lenalidomide-dexamethasone combination, with selection of different subclones at both first and second relapses.37 The only way to answer this question would be to systematically analyze the diagnostic and first relapse specimens in a homogeneously treated cohort of patients.
Subclonal evolution. Two different types of subclonal evolution can be observed: a “linear” evolution with accumulation of genetic events (A) and a “branching” evolution with early divergence of subclones with different mutations, which are differentially selected during evolution (B).
Subclonal evolution. Two different types of subclonal evolution can be observed: a “linear” evolution with accumulation of genetic events (A) and a “branching” evolution with early divergence of subclones with different mutations, which are differentially selected during evolution (B).
Multiple myeloma: a single or multiple disease?
Whatever way myeloma is analyzed (clinically, cytogenetically, or molecularly), it is a very heterogeneous disease. The question is whether this heterogeneity hides several subentities of multiple myelomas. Actually, there is at least 1 precedent in hematology with the molecular dissection of diffuse large B-cell lymphomas (DLBCLs). Whereas this type of lymphoma was identified as a single disease based on pathological features, the molecular analyses using gene expression profiling (GEP) dissected the DLBCL in at least 3 subentities, the activated B-cell type, the germinal center type, and the mediastinal type, mainly defined by differences in cells of origin.39,40 This finding was not only intellectual because it was later shown that these types of DLBCL responded differently to various therapeutic schemas. The question has been addressed in MM by several groups, and so far, we have to recognize that this goal has not been reached. Using GEP and nonsupervised analyses, 3 reports identified subgroups mostly driven by chromosomal aberrations.41-43 The first report identified 8 different subgroups, mainly based on the cyclin D gene expression and on the different 14q32 recurrent translocations.41 This molecular classification was refined in 2006, identifying 7 subclasses of myelomas.42 A first class was defined by the translocation t(4;14), identified by overexpression of the MMSET and/or FGFR3 genes. The second one was defined by the upregulation of one of the MAF genes, related to the translocations t(14;16) or t(14;20). Cases with CCND1 or CCND3 upregulation [because of the translocations t(11;14) or t(6;14)] clustered in 2 different groups, named CD1 and CD2. The CD2 group was characterized by CD20 expression. The fifth group was characterized by hyperdiploidy. The 2 last groups were characterized by a low incidence of bone disease, according to a low dickkopf WNT signaling pathway inhibitor 1 expression, whereas the last group was characterized by high expression of genes involved in proliferation. This molecular classification has been partially confirmed by a study from the Haemato-Oncology Foundation for Adults in the Netherlands group.43 The “low bone disease” group was not confirmed. In contrast, 3 other groups were identified: 1 group enriched by “myeloid” genes (that could be related to plasma cell sorting problems); 1 group characterized by overexpression of cancer testis antigen genes; and, finally, a group defined by overexpression of positive regulators of the nuclear factor κB pathway, which seems to be important in the MM oncogenesis. Whether these subclasses define true MM subentities is not yet demonstrated. Although various GEP-based signatures have been reported to identify high-risk disease and predict prognosis, a recent study suggests inability of GEP to predict response in myeloma.44
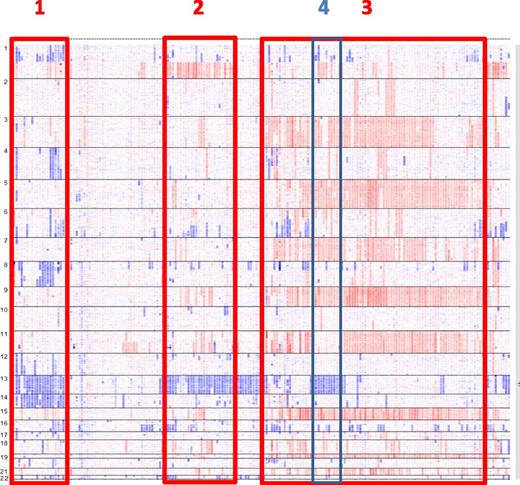
Another way to approach this question is to use CGH/SNP-array analyses.30,45 Based on these analyses, several subgroups of patients can be individualized. Apart from the hyperdiploid group (which appears also heterogeneous based on the trisomy combinations), some subgroups can be identified, such as a group characterized by monosomies 13 and 14, with frequent 1p deletions, or another one characterized by monosomy 13 and 1q gains (Figure 4). Whether these subgroups represent different myeloma entities remains to be explored.
MM subentities defined by SNP array. In this picture of 192 patients with MM at diagnosis, several subgroups can be identified: monosomies 13 and 14 and frequent 1p deletions (1); monosomies 13 and 1q gains (2); hyperdiploidy (3); and within hyperdiploidy, patients with monosomies 13 and lack of trisomy 11 (4). Adapted from Avet-Loiseau et al30 with permission.
MM subentities defined by SNP array. In this picture of 192 patients with MM at diagnosis, several subgroups can be identified: monosomies 13 and 14 and frequent 1p deletions (1); monosomies 13 and 1q gains (2); hyperdiploidy (3); and within hyperdiploidy, patients with monosomies 13 and lack of trisomy 11 (4). Adapted from Avet-Loiseau et al30 with permission.
Finally, this question has been addressed by using massively parallel sequencing. This technique has been very successful in several tumor types to identify unique gene mutations driving the oncogenesis of the disease. For instance, in hematology, this approach identified 2 malignancies that display an almost constant single gene mutation (ie, MYD88 in Waldenström macroglobulinemia46 and V660E BRAF mutation in hairy cell leukemia47 ). Three major papers addressed the issue of the mutational landscape of MM using next-generation sequencing (NGS).48-50 The first study analyzed 38 patients at different stages (diagnosis, relapse) and showed that the distribution of mutations was widely variable between patients, confirming the large heterogeneity observed at the clinical or biological level. No common frequent mutation was observed.48 The median number of mutations per genome was ∼55 to 60, with a very large range (21-488). When compared with other tumors, MM is in the middle, between low-mutated tumors such as leukemias and highly mutated carcinogen-induced tumors.49 Furthermore, all 3 reports showed that only a few mutations are recurrent, such as NRAS and KRAS mutations. Some other mutations are observed in a few percent of the patients, but <30 genes are recurrently mutated in at least 5% of the patients. Of note, many of these mutations are observed only in subclones, including genes supposed to act as drivers (NRAS, KRAS, BRAF, for instance).49,50 These data have important implications for the treatment of patients with specific B-Raf proto-oncogene or MAP kinase-ERK kinase inhibitors. Their therapeutic effect should be maximal only in cases where these driver mutations are present in all tumor cells. Although some of these mutations may provide sensitivity to specific targeted agents, such as vemurafenib in patients with BRAF V600E mutation in myeloma, the functional impact of some of these mutations remains ill-defined. Thus a larger mutation-directed therapeutic intervention study will need to be performed to define their impact. Moreover, innovative methods need to be developed to measure response, especially response that may only be directed at the clones carrying the mutation.51 These data are even strengthened by a recent observation that these supposed driver mutations are not always expressed at the RNA level, highlighting the need for future analyses at the protein level.52 Some of these sequencing studies have also further defined the subclonal content and their evolution over time as well as defined potential mutational signatures used during progression. These results have raised questions about what ongoing mechanisms may be operative during clonal evolution. An important need now is to define the role of some of the known DNA repair mechanisms in inducing clonal change.53
Clinical implications
As demonstrated in most hematologic malignancies, genetic changes play a major role in prognostication in MM. However, in contrast to leukemias, no “good-risk” abnormalities have been described so far. Among the high-risk chromosomal abnormalities, the most powerful ones are del(17p), t(4;14), and del(1p32).54-59 These abnormalities impact significantly both progression free survival and overall survival. Of note, these high-risk factors do not impact response to therapy, including the del(17p), raising questions about the impact of del(17p) on TP53. Mutational studies did show that TP53 mutations are more frequent in patients with del(17p), but only in 30% to 40% of them.60 Other chromosomal changes do impact survival, such as 1q gains,61 del(12p),30 or t(14;16). However, their prognostic significance is either low (1q gains) or not confirmed in all studies [del(12p) and t(14;16)]. Regarding other recurrent chromosomal changes, such as t(11;14) or hyperdiploidy, they are associated with a standard risk, even though hyperdiploidy is probably heterogeneous and may contain some good-risk combinations. So far, the identification of good-risk patients is essentially based on the absence of high-risk genetic features, associated with a low β2-microglobulin level.62 In an ongoing SNP-array study, we showed that genetic abnormalities represent the major prognostic value, representing 75% of the overall survival prediction. Preliminary analyses of NGS data did not suggest that mutations display specific prognostic value, although larger systematic studies are warranted to clarify this point, especially to detect the role of subclonal variants of a number of mutations.
These DNA-based approaches and several studies analyzing GEP have shown the ability to predict patient outcome.42,43,63 Of note, all these models are defined by various sets of genes with a very limited number of genes in common. Whether these differences reflect differences in the therapeutic schemas used to define the models or just biomarkers is currently unknown. However, all these models are able to predict patient survival, even though a recent study did show that GEP was not able to predict response.44 Similar findings were previously reported with FISH, with high-risk patients presenting similar response rates as standard-risk patients. Overall, efforts by our group and others have defined the role of FISH and GEP in developing a prognostic model, with limited initial contribution from NGS studies. Further understanding of clonality and clonal evolution may improve these models in future
Conclusion
Despite the use of the most promising genomic tools (GEP, SNP array, and NGS), MM remains a very heterogeneous disease, with no unique common mutation. The NGS studies in the past 5 years have characterized the suclonality concept. Even though NGS data can be considered disappointing because they do not show common mutations that could define subentities, they are nevertheless important because they confirm the wide molecular heterogeneity of the disease and the frequent occurrence of some supposedly “driver” mutations only in subclones. This finding is important not only for our understanding of the biology of these genes in MM, but also for the hope of targeted therapies. It is clear that improvement in our understanding has been incremental and not exponential with successive utilization of various technologies and the molecular dissection of the disease, especially in the demonstration of suclonality. The objectives of future studies will be to analyze the 100 to 200 recurrent mutations (clonal or subclonal), in homogeneously treated cohorts of patients, in order to understand their significance in the evolution of MM, but also to learn how to deal with them in order to propose the best treatment to patients.
Acknowledgments
This work was partially supported by the CAPTOR program, and a National Institutes of Health, National Cancer Institute PO1 grant (CA 100707-12).
Authorship
Contribution: All authors wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hervé Avet-Loiseau, IUCT-Oncopole, CHU, Toulouse, 31059, France; e-mail: avet-loiseau.h@chu-toulouse.fr.