Abstract
Understanding how tumor cells fundamentally alter their identity is critical to identify specific vulnerabilities for use in precision medicine. In B-cell malignancy, knowledge of genetic changes has resulted in great gains in our understanding of the biology of tumor cells, impacting diagnosis, prognosis, and treatment. Despite this knowledge, much remains to be explained as genetic events do not completely explain clinical behavior and outcomes. Many patients lack recurrent driver mutations, and said drivers can persist in nonmalignant cells of healthy individuals remaining cancer-free for decades. Epigenetics has emerged as a valuable avenue to further explain tumor phenotypes. The epigenetic landscape is the software that powers and stabilizes cellular identity by abridging a broad genome into the essential information required per cell. A genome-level view of B-cell malignancies reveals complex but recurrent epigenetic patterns that define tumor types and subtypes, permitting high-resolution classification and novel insight into tumor-specific mechanisms. Epigenetic alterations are guided by distinct cellular processes, such as polycomb-based silencing, transcription, signaling pathways, and transcription factor activity, and involve B-cell-specific aspects, such as activation-induced cytidine deaminase activity and germinal center–specific events. Armed with a detailed knowledge of the epigenetic events that occur across the spectrum of B-cell differentiation, B-cell tumor–specific aberrations can be detected with improved accuracy and serve as a model for identification of tumor-specific events in cancer. Insight gained through recent efforts may prove valuable in guiding the use of both epigenetic- and nonepigenetic-based therapies.
Introduction
Epigenetic modifications are defined as heritable changes to the chemistry of DNA (without alteration to the underlying nucleotide sequence) or modification to the structure or proteins that compose chromatin. The purpose of these modifications is to create and maintain a stable, global gene expression program of carefully calibrated expression states and tuned responses to upstream signals. Collectively, the global pattern of epigenetic modifications is the software that uniquely defines the applications performed by a given cell type and that ultimately becomes corrupted in cancer. A multitude of epigenetic modifications work in concert to regulate normal and aberrant gene behavior; this review will primarily focus on DNA methylation (5′-methylcytosine, 5mC) as it is the most broadly studied epigenetic modification in B-cell tumors. DNA methylation occurs mainly at cytosine guanine dinucleotide (CpG) dinucleotides in mammals, and its presence and function are governed by a concert of DNA methyltransferases (writers), 5mC binding proteins (readers), and a series of chemical conversions that contribute to 5mC demethylation involving ten-eleven translocation methylcytosine dioxygenase (TET) and thymine-DNA glycosylase (TDG) enzymes1 (erasers). Cytosine demethylation can also occur passively by failure to methylate nascent DNA during cell replication. DNA methylation levels generally correlate with CpG content, with low levels in CpG-enriched regions (CpG islands) and higher methylation elsewhere. A multitude of gene-based studies have reported that promoter DNA methylation is inversely correlated with gene expression.2 Although this correlation still holds true for some genes, recent reports at the genome scale suggest that such association is less clear than previously appreciated and that the functions of DNA methylation can be different in distinct genomic contexts, such as promoters, enhancers, gene bodies, and heterochromatin/repetitive elements.3 Furthermore, understanding the function of DNA methylation requires an integrative analysis with histone marks that collectively designate chromatin into different chromatin states.4 This analysis reveals that 5mC levels are low in active or poised regulatory elements and high in transcribed and heterochromatic regions.5 Beyond the context-dependent function of DNA methylation, cells passively accumulate DNA methylation changes in repressed regions without any apparent functional impact and can be used as a mitotic clock.6 Thus, DNA methylation can be used to infer basic properties of the local chromatin state as well as the developmental and proliferative cellular history. Conveniently, this information is covalently archived on the DNA molecule itself and thus accessible without the additional caveats of protein and chromatin-based analysis.
All malignancies studied to date exhibit altered epigenetic patterns, and B-cell malignancies are no exception. The emerging picture from genome-wide DNA methylation data illustrates a highly abundant landscape of alterations in B-cell tumor types and subtypes (Figure 1). In this review, we will address the following: (1) What alterations to DNA methylation occur in B-cell malignancies? (2) How and when do changes occur? (3) What changes are more likely to be important for altered tumor identity? (4) Can this information be used to guide clinical practice?
Illustration of DNA methylation alterations observed in B-cell malignancies. Several types of tumor-specific changes have been observed in genome-wide DNA methylation studies and are commonly associated with chromatin states in normal B cells. Global hypomethylation is enriched in intergenic regions associated with heterochromatin and quiescent chromatin states. Additional targeted hypomethylation occurs across the methylome of malignant B cells, which is enriched in active gene bodies and the binding sites of overexpressed transcription factors (TFs) and in regions immediately downstream of promoters. Hypermethylation of enhancer regions resulting in enhancer decommissioning is also enriched in gene bodies and correlates with loss of TF expression. Finally, all B-cell malignancies demonstrate hypermethylation of promoters and other sequences associated with polycomb repression in precursory normal cells.
Illustration of DNA methylation alterations observed in B-cell malignancies. Several types of tumor-specific changes have been observed in genome-wide DNA methylation studies and are commonly associated with chromatin states in normal B cells. Global hypomethylation is enriched in intergenic regions associated with heterochromatin and quiescent chromatin states. Additional targeted hypomethylation occurs across the methylome of malignant B cells, which is enriched in active gene bodies and the binding sites of overexpressed transcription factors (TFs) and in regions immediately downstream of promoters. Hypermethylation of enhancer regions resulting in enhancer decommissioning is also enriched in gene bodies and correlates with loss of TF expression. Finally, all B-cell malignancies demonstrate hypermethylation of promoters and other sequences associated with polycomb repression in precursory normal cells.
To fully harness the potential for epigenetics in cancer therapy, obtaining a comprehensive, genome-wide picture is vital to clearly identify underlying pathogenic events and to ascertain the most useful diagnostic/predictive information. Studies prior to the widespread incorporation of genome-wide profiling technologies mainly focused on promoter methylation of candidate genes, revealing numerous examples of gene-specific alterations in DNA methylation in a variety of B-cell tumors. Here we will focus mainly on emerging concepts learned from recent studies investigating DNA methylation using high-resolution genome-wide approaches (Illumina 450K arrays and next-generation sequencing) in relatively large patient cohorts to provide a comprehensive genome-wide view of various B-cell malignancies. Important to the understanding of B-cell tumors are data provided from parallel studies of normal B-cell differentiation, revealing a remarkable degree of change in normal cells (recently reviewed by Martin-Subero and Oakes7 ). Briefly, these modifications involve highly orchestrated focal changes to promoters and regulatory regions, as well as reduction in global DNA methylation levels with increasing maturation. We will elaborate subsequently on how this process underlies the biology of individual tumor types and subtypes.8,9
Global DNA methylation loss is highly prevalent in B-cell malignancies
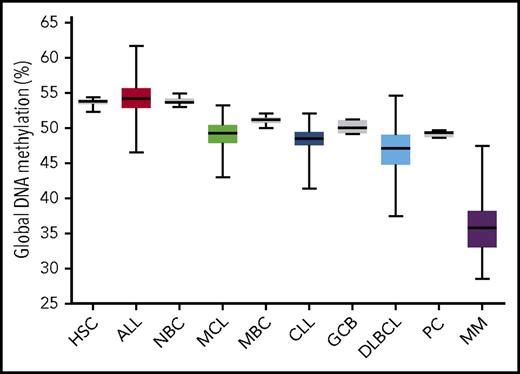
An immediate obvious feature of DNA methylation in B-cell malignancies is the overall global reduction in DNA methylation levels observed in most tumor types investigated, with the exception of acute lymphocytic leukemia (ALL) (Figure 2).8,10-14 The majority of this loss can be attributed to partial loss of methylation in heterochromatic, intergenic DNA sequences. Global methylation loss is not unique to B-cell tumors per se, as it is also observed in Epstein-Barr virus–transformed lymphoblastoid cell lines15,16 and is observed in some solid tumors.17,18 Interestingly, the general degree of global hypomethylation in B-cell tumors scales in parallel with the methylation level of each tumor type’s inferred founding normal cell type (ie, ALL shows the least hypomethylation and is the most phenotypically immature, similar to pre–B cells, whereas multiple myeloma (MM) exhibits the most hypomethylation and plasma cells naturally retain the lowest methylation levels). Reduction of tumor global methylation levels relative to the normal state likely results from enhanced cell proliferation following transformation. In CLL patients, universal loss of DNA methylation occurs concurrently with local disorder of methylation patterns.19 The broad nature of global methylation loss suggests that passive mechanisms are likely the primary cause; however, active demethylation mechanisms may also be involved. In general, global hypomethylation has been linked to genetic instability20 ; however, as normal mature B cells also lose methylation and heavily hypomethylated B-cell tumors are not necessarily more genetically unstable, global hypomethylation per se does not seem to cause instability. The impact on B-cell tumors is currently unclear.
Global DNA methylation levels in normal and malignant B cells. Illumina 450K array data were obtained from multiple studies of normal and tumor cells.8-11,13,38,47,84 Normal/tumor pairs are ordered according to degree of normal B-cell differentiation. Illumina array data were uniformly normalized; percent global methylation is represented by the average array β value × 100. Normal cells illustrated in gray. Boxes represent interquartile range, and whiskers represent minimum/maximum values. CLL chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B cell; HSC, CD34+ hematopoietic stem and progenitor cell; MBC, memory B cell; MCL, mantle cell lymphoma; MM, multiple myeloma; NBC, naïve B cell; PC, bone marrow plasma cell.
Global DNA methylation levels in normal and malignant B cells. Illumina 450K array data were obtained from multiple studies of normal and tumor cells.8-11,13,38,47,84 Normal/tumor pairs are ordered according to degree of normal B-cell differentiation. Illumina array data were uniformly normalized; percent global methylation is represented by the average array β value × 100. Normal cells illustrated in gray. Boxes represent interquartile range, and whiskers represent minimum/maximum values. CLL chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B cell; HSC, CD34+ hematopoietic stem and progenitor cell; MBC, memory B cell; MCL, mantle cell lymphoma; MM, multiple myeloma; NBC, naïve B cell; PC, bone marrow plasma cell.
DNA hypermethylation is guided by polycomb repression
DNA hypermethylation in B-cell tumors is highly associated with regions known to exhibit polycomb repression in normal precursor cells. Polycomb repression is a mechanism of gene silencing based on trimethylation of histone H3, lysine 27 (H3K27me3) by the EZH2 methyltrasferase of the polycomb repression complex 2 (PRC2). Once modified, the complex recruits PRC1 and other corepressive factors for stable silencing. Polycomb repression is used by developing cells to suppress alternate lineage-specifying gene expression programs and can involve simultaneous repressive H3K27me3 and active H3K4me3 modifications at promoters, termed a “bivalent” (poised) chromatin state.21 Polycomb repressed/bivalent regions in normal cells are generally devoid of DNA methylation,22 but at some loci in tumor cells, H3K27me3 is replaced with DNA methylation in a process termed “epigenetic switching.”23 In B-cell malignancies, polycomb-marked regions display the highest enrichment for gain of DNA methylation10-13 ; however, insufficient matched H3K27me3 chromatin immunoprecipitation–sequencing data are currently available to document concurrent H3K27me3 loss and switching. Nonetheless, an extensive “polycomb repression-associated DNA methylator phenotype” (PRAMP) is observed across B-cell malignancies as well as in many other cancer types.24-26 Although the exact cause is unknown, it may result from excessive cell proliferation, as forcing excessive proliferation of normal cells and disruption of the key cell cycle regulator, CDKN2A, induces PRAMP.27,28 Indeed, hypermethylation of polycomb regions is associated with genetic mutations in other tumors that induce mitogenic signaling, such as BRAF.29 Furthermore, PRAMP is associated with advanced chronological age and is a component of a molecular aging signature.30 Although PRAMP is broadly observed in tumor genomes and commonly involves promoters, the tangible impact on gene expression is unclear as polycomb-repressed genes often already display low/absent expression in normal precursor cells. However, PRAMP is associated with a further reduction of expression,31 and conversely, occasionally genes display increased expression in tumors.12,28,32 Although PRAMP-associated changes are likely a consequence of proliferation, PRAMP may provide a mechanism to further stabilize the repression of genes that would be undesirable to the tumor cell if reexpresssed.23
TFs direct focal methylation changes at enhancers and gene bodies
Large-scale epigenomic profiling of multiple tissue and tumor types has emphasized that the vast majority of DNA methylation and other epigenetic modifications occur outside of promoters, highlighting the role that these other functional DNA elements play in controlling gene expression programs.5,33,34 When enhancers are activated (usually associated with DNA hypomethylation and gain of the active histone marks H3K27ac and H3K4me1), they can impact gene expression levels, enable different responses to intrinsic signaling, and have other regulatory roles such as RNA splicing.35,36 Various DNA methylation profiling studies in ALL, CLL, MCL, Burkitt lymphoma (BL), follicular lymphoma (FL), and MM have revealed a concentration of altered DNA methylation at enhancers.10-13,37 B-cell malignancies mostly exhibit tumor-specific hypomethylation at enhancer loci, with the exception of MM, which displays enhancer hypermethylation and decommissioning.11 In MCL, de novo hypomethylation and activation of a distant enhancer is associated with overexpression of the SOX11 oncogene through a long-range three-dimensional interaction.13 There is also a predisposition for focal alterations of DNA methylation to occur in intragenic (gene body/intronic) sequences as opposed to intergenic regions.10,12,13,38 MM exhibits hypermethylation of enhancers in intragenic regions.11 Intragenic sequences normally display high levels of DNA methylation, which can further increase during B-cell maturation.9 Intragenic methylation levels also proportionally increase with transcription rates,12,39 and actively transcribed genes display distinct histone modifications, such as H3K36me3. Recent work has linked H3K36me3 to the specific recruitment of DNMT3B and subsequent DNA hypermethylation of transcribed regions,40-42 suppressing alternative intragenic transcriptional initiation during differentiation.43 Methylome and RNA sequencing in BL and FL revealed that DNA methylation changes within intragenic regions were highly correlated with gene expression, the most of any chromatin compartment.12 The enrichment of DNA methylation changes in intragenic vs intergenic DNA suggests that either intragenic methylation is intrinsically more unstable in tumor cells or that these changes are selected during tumorigenic processes.
B cells employ a specific set of TFs to drive differentiation. DNA hypomethylation is highly enriched at enhancer loci bound by these TFs in maturing B cells.8,9,44,45 In turn, tumor cells employ TFs to modify epigenetic states. ALL is characterized by distinct genetic subtypes, many of which are defined by a single fusion gene involving 1 or more TFs.46 These subtypes demonstrate remarkably distinct methylation patterns tightly associated with the presence of the TF fusion,31,38 strongly suggesting that functional disruption of the TF is responsible for aberrant patterning. Similarly, TF fusions partner with specific methylation patterns in acute myeloid leukemia (AML).47 In FL and BL, tumor- and subtype-specific DNA methylation differences were enriched at several transcription factor binding sites (TFBSs), including hypomethylation of TCF3/E2A and hypermethylation of STAT, YY1, MEF2, and BCL3 binding sites.12 Altered TFBS methylation correlates with correspondingly altered TF expression, downstream target gene/pathway activity, and in some cases, altered TF gene methylation. Enhancer hypermethylation in MM correlated with corresponding decreased TF expression.11 In CLL, tumor-specific hypomethylation is enriched in the binding sites for TFs involved in B-cell development, such as TCF3 and PU.1/SPIB, and also some TFs involved in B-cell activation, such as NFAT and EGR.9 Hypomethylation of the latter 2 TFBSs was enriched in the more aggressive subtype of CLL,9 which also displays stronger B cell receptor (BCR) activation and signaling.48 As the activation of NFAT and EGR TFs occurs downstream of BCR stimulation (via calcium influx and MAPK induction, respectively), it suggests that remodeling of the epigenetic landscape in CLL is potentially linked to hyperactive BCR activity.
Targeted active demethylation
Whereas loss of global DNA methylation of heterochromatic regions is thought to occur through a passive, replication-dependent process, targeted hypomethylation and enhancer generation may occur actively. In vitro stimulation of naïve B cells caused early, targeted demethylation that was not dependent on the degree of proliferation.9 Similarly, Caron and colleagues found that DNA demethylation during plasma cell development in vitro occurred in response to differentiating stimuli and was not principally linked to proliferation.49 This study also noted an association of 5-hydroxymethylcytosine at regions undergoing targeted demethylation and activation, implicating an active demethylation pathway. Although it is currently an open question how certain TF binding events may result in targeted DNA methylation, recruitment of TET family enzymes may be involved. Indeed, TET2 was found to directly interact with EBF1,50 an essential factor for B-cell development, which is highly enriched at focally hypomethylated sequences in mature B cells.9,51 Another possibility involves active deamination of 5mC by the activation-induced cytidine deaminase (AID)/APOBEC family cytosine deaminases. An early study reported enrichment of AID motifs and SHM-targeted regions in hypomethylated sequences in GCBs.52 Recent work in AID knock-out mice demonstrated an attenuated germinal center (GC) DNA hypomethylation signature.53 Although AIDs role in DNA demethylation is controversial,54,55 its role is further supported by studies of active demethylation in the germ line and development.56 Although AID has been less studied in B-cell malignancies, overexpression in mice causes more aggressive lymphomas with more diverse intertumor methylation heterogeneity.57 AID is found to be overexpressed and active in several B-cell tumor types,57,58 warranting study of AID-dependent demethylation more broadly in B-cell malignancies.
When do changes occur?
Tumor-specific DNA methylation changes in B-cell malignancies can potentially arise at many points throughout the natural history of a tumor cell population, from the establishment of the founder cell through to relapse disease stages. The most longitudinal information is currently available for CLL, with several groups reporting genome-wide DNA methylation data at various time points. Remarkably, DNA methylation patterns are generally highly stable across many years of disease course (including spanning treatment), rarely displaying alterations involving >2% of observed CpGs.59-62 A similar degree of stability was observed in FL.63 Targeted deep sequencing measuring DNA methylation patterns on individual reads (to infer intratumor heterogeneity of methylation patterns) revealed the existence of highly clonal, allele-specific patterns of DNA methylation in stable CLL cases.59 This suggests that most of the tumor-specific DNA methylation observed in samples obtained in clinical disease phases is established prior to, or at the time of, formation of the founder cell.
Although a greater degree of DNA methylation stability was observed in many CLL and FL cases, this is likely not a broad characteristic of all B-cell malignancies. Malignant B cells may arise from a broad range of differentiating premalignant B-cell subpopulations.8 Phenotypically, normal pre-GC naïve B cells are poised to enact changes upon antigen interaction, GCBs are actively evolving, and post-GC memory B cells are tasked to stably maintain their postevolution phenotype. Thus, if tumors arising from pre-GC and GC stages retain a component of these attributes, they may have the potential to generate greater DNA methylation instability, perhaps through the expression of AID.64 CLL cases that exhibited a higher degree of DNA methylation changes were more commonly of the IGHV-unmutated, GC-independent subtype.59 In DLBCL and ALL, evolution of DNA methylation was observed in the majority of cases between diagnosis and relapse,38,65 which may relate to the underlying developmental, evolving phenotypes of the respective cell of origin. Although much work remains to be done in the longitudinal evaluation of epigenomic features in B-cell malignancies, these early data suggest that stability and evolution of DNA methylation is associated with more indolent and aggressive disease, respectively.
Identification of truly aberrant epigenetic changes from a dynamic spectrum of precursors
Unlike somatic genetic alterations that can be identified relative to the germline sequence, deciphering alterations in epigenetic profiles presents the additional challenge of cell-type variability. As maturation stages of normal B-cell types carry distinct epigenetic patterns, the epigenetic landscape in any given tumor cell is the combination of that which is acquired through normal developmental programming and tumor-specific events (Figure 3). Thus, the identification of true tumor-specific alterations necessitates careful selection of an appropriate normal reference epigenome. This challenge is further compounded by the fact that tumors generally arise from a single founder cell (which can be elegantly confirmed in B-cell malignancies via clonal immunoglobulin rearrangement). This unpredictable variation in any given normal cell when blended with tumor-specific event(s) creates an “epigenetic uncertainty principle,” as is it is impossible to know a priori the normal epigenetic state in the founder cell prior to transformation.
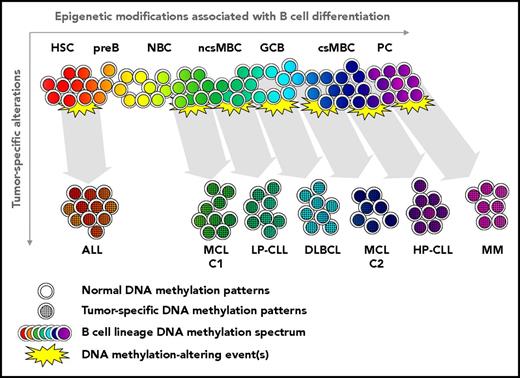
Relationship of epigenetic alterations in normal B-cell and tumor development. Substantial genome-wide epigenetic modification occurs in sequential normal B-cell subpopulations, creating a continuum of epigenetic states involving targeted modification of promoters and enhancers, as well as global changes. Transforming event(s) (genetic, epigenetic, and/or others) are associated with alteration of the epigenetic state occur in a founder cell. The resulting tumor epigenetic pattern is a composite of the original B-cell and tumor-specific events. Subtypes of the same B-cell malignancy may derive from different stages of normal B-cell differentiation, as documented in MCL13 and CLL.9,10 Tumors exhibit an accelerated degree of differentiation relative to inferred normal cell of origin (including substantial global DNA methylation loss), permanently obscuring knowledge of the precise original phenotype. MCL-C1 and -C2, mantle cell lymphoma cluster 1 and 2, respectively; LP- and HP-CLL, low- and high-programmed CLL, respectively; ncsMBC and csMBC, non-class-switched and class-switched memory B cells, respectively.
Relationship of epigenetic alterations in normal B-cell and tumor development. Substantial genome-wide epigenetic modification occurs in sequential normal B-cell subpopulations, creating a continuum of epigenetic states involving targeted modification of promoters and enhancers, as well as global changes. Transforming event(s) (genetic, epigenetic, and/or others) are associated with alteration of the epigenetic state occur in a founder cell. The resulting tumor epigenetic pattern is a composite of the original B-cell and tumor-specific events. Subtypes of the same B-cell malignancy may derive from different stages of normal B-cell differentiation, as documented in MCL13 and CLL.9,10 Tumors exhibit an accelerated degree of differentiation relative to inferred normal cell of origin (including substantial global DNA methylation loss), permanently obscuring knowledge of the precise original phenotype. MCL-C1 and -C2, mantle cell lymphoma cluster 1 and 2, respectively; LP- and HP-CLL, low- and high-programmed CLL, respectively; ncsMBC and csMBC, non-class-switched and class-switched memory B cells, respectively.
Finding a way to mitigate this uncertainty is vital in deducing true tumor-specific epigenetic alterations. The uncertainty can be addressed in part by understanding the range of changes that occur within the appropriate normal cell lineage(s), and then tumor-specific events can be identified as those that occur outside the defined normal range. In addition, as with mutations, a significant proportion of alterations in a given individual tumor are likely random passenger events, necessitating the study of sufficient numbers of patients to define highly recurrent epigenetic alterations. Finally, contamination of tumor samples with varying amounts and types of normal cells can be solved by computational methods that can deconvolute and purify the tumor cell methylation signal in silico.13,66 This combined approach has been applied in CLL and MCL where DNA methylation is compared relative to dynamic changes occurring in the normal B-cell lineage.9,13 These analyses revealed that 65% to 80% of the changes observed in CLL and MCL samples also occur during normal B-cell development.9,13 Similar results were obtained for ALL and DLBCL.8 Indeed, an analysis of a collection of genes previously found to display hypermethylation in CLL67 revealed that of 29/30 genes, the methylation change during normal B-cell differentiation was greater or equal to what was acquired in CLL. Mistaken hypermethylation was mainly because of memory B cells naturally exhibiting PRAMP, whereas naïve B cells do not, which compose the majority of B cells in unselected normal controls. Despite uncertainty surrounding if tumor-specific methylation events were acquired prior to or after transformation, the fact that these specific epigenetic states exist in the normal memory B-cell pool (which generally subsists for decades) forgo these alterations as being directly involved in tumorigenesis. The results of these studies highlight the need for caution in the interpretation of tumor epigenetic profiles and warrant the application of this approach to other tumor types.
Focusing on genuine tumor-specific DNA methylation changes in CLL revealed that most regions that were hypermethylated resulted from the failure to achieve the DNA methylation state that occurs in normal B-cell maturation (ie, a blockage in directed hypomethylation).9 These regions were highly enriched for binding sites of the TFs EBF1, BATF/AP-1, RUNX3, and others. These TFs are known to be involved in normal B-cell differentiation, suggesting that selective blockage of these pathways is involved in CLL pathogenesis. Combining findings of genuine hypomethylation being associated with other TFs, such as EGR and NFAT, indicates an imbalance of the activities of key TFs involved in normal B-cell maturation, and activation is a significant driver of epigenetic alterations in CLL. Further work will need to be done to uncover which pathways and aberrant downstream epigenetic events directly influence tumorigenesis.
Genetics vs epigenetics: who is driving who?
A wealth of data surrounding somatic mutations and epigenetic alterations in B-cell malignancies begs the question of the relationship between these events. In ALL, chromosomal rearrangements are highly correlated with methylation subtypes,38 suggesting a genetic basis for these epigenetic changes. A genetic origin is further supported by AML data, which also involve single-nucleotide mutations, such as IDH1/2, NPM1, and DNMT3A.47,68 However, the association of genetic/epigenetic changes are less clear in more mature B-cell tumor types. The most comprehensive data currently available are for CLL, where certain genetic events are nonrandomly enriched across distinct DNA methylation subtypes. For example, recurrent deletions of chromosomes 11q and 14q, as well as mutations in NOTCH1, SF3B1, and EGR2 were biased among methylation subtypes.9,10 Additionally, overall mutation rate associates methylation subtype.69 In MCL, methylation subtypes display a differential enrichment of overall copy-number alterations.13 However, these associations found in non-ALL/AML fundamentally differ, as genetic events are merely enriched and do not generally define the subtype-specific methylation pattern. Methylation subtype patterns in CLL are predominantly derived from the broad methylation signature present in the founder cell. As the majority of mutations in CLL exhibit subclonal frequencies,69,70 the overall tumor methylation pattern that defines the epigenetic subtypes precedes the acquisition of mutation(s) in most instances. Intriguingly, this implies that the tumor epigenetic state may regulate which mutations provide an advantageous pairing. Indeed, pan-cancer data clearly show that the occurrence of recurrent genetic mutations is nonrandom across tumor types, highlighting the importance of cellular context governing mutation acquisition. In CLL, construction of individual tumor phylogenies has revealed multiple distinct mutations occurring in the same gene in separate evolutionary branches,71 suggesting that a particular root phenotype strongly benefits from an explicit recurrent mutation. Furthermore, genetic and DNA methylation changes were found to co-occur in FL and CLL.59,63 Overall, the data support a role for a mutual relationship between genetic and epigenetic events in tumors, depending greatly on the type of mutation, when it arises in tumor development (ie, clonal vs subclonal), and differentiation stage/cellular context.
Prospective clinical implications
The epigenomes of B-cell malignancies are rich in information that recount the normal and pathogenic events that shaped the identity of an individual tumor, providing high-resolution data for diagnostic and subtype classification and to potentially reveal exploitable vulnerabilities. DNA methylation is highly stable in many B-cell tumor types, permitting a favorable setting for biomarker development, as measurement is thus not dependent on sample timing or impacted by sample handling,72 and furthermore can be analyzed from liquid biopsies.73 Early evidence of this has come from the study of the ZAP70 locus in CLL, where this powerful cytometry-based prognostic marker was markedly improved by measuring promoter-proximal methylation.74,75 As expression of ZAP70 benefits CLL cells,76 it appears paradoxical that a mark of gene regulation would be more impactful than the protein level itself. As cell state and/or technical handling of samples affects protein stability,77 methylation marks may more accurately represent the general propensity of a gene to be expressed over the course of disease and in different anatomical locations. Indeed, although CLL cells exhibit dramatically different transcriptomes depending on being located in lymph nodes or in peripheral blood,78 genome-wide methylation patterns are remarkably similar.79 A second, nonmutually exclusive explanation for the clinical value of ZAP70 methylation is that the locus is just one part of a larger global signature associated with prognosis.
Unsupervised analysis of genome-wide DNA methylation patterns has facilitated the data-driven identification of novel epigenetic subgroups within B-cell malignancies. In CLL, broad, genome-wide DNA methylation patterns (which include ZAP70) define 3 distinct tumor subtypes.9,10,60 These subtypes differ primarily in the degree of normal maturation achieved by the founder cell and thus retain markings of normal B cells in addition to tumor-specific changes. Importantly, each subtype displays distinct clinical outcomes across multiple cohorts and retains independent prognostic power when traditional prognostic markers are considered.60 Methylation subgroups displaying a less mature (GC-independent) pattern are associated with poorer outcomes, and inferior outcomes are also associated with less maturity within subtypes.9 Unsupervised analysis of MCL reveals 2 methylation subtypes with clinical impact.13 Interestingly, within MCL subtype designations, the greater the total number of DNA methylation changes (developmental and tumor-specific) associates with inferior outcomes. In ALL, subtype-specific differential methylation divided patients into different risk groups for relapse-free survival.38 Arribas and colleagues identified 2 splenic marginal zone lymphoma subtypes, 1 displaying extreme PRAMP and associating with poor outcome.80 Genome-wide methylation-based classification is a novel and promising way to classify tumors that may add to genetic classification in prognostic index models, especially in tumor types that exhibit low frequencies of recurrent mutations.
In the future, epigenetic information may be used for matching patients to individualized therapy. For example, the subset of CLL patients that retain greater BCR signaling capacity experience greater responses to BCR inhibition.81 These patients exhibit a DNA methylation signature of NFAT hyperactivity,9 suggesting that identification of this signature in other tumor subtypes across the B-cell malignancy spectrum may pinpoint patients with a similar therapeutic benefit. Interestingly, several B-cell malignancies exhibit a high frequency of genetic alterations in components of the epigenetic machinery, including key histone acetyl- and methyltrasferases.82 Tumor-specific epigenetic patterns may integrate with genetic data to guide the use of emerging epigenetic therapies. Beyond DNA methylation, current efforts are being focused on mapping other marks such as histone modifications, chromatin accessibility, and three-dimensional chromatin structure83 ; undoubtedly integrating these analyses will provide new understanding of the biology of B-cell tumors, with novel clinical applications. Intratumor epigenetic heterogeneity and subclonal architecture uncovered through employing single-cell technologies may provide important prognostic and response monitoring data.
Acknowledgments
The authors thank their primary funding sources for studies on epigenomics.
C.C.O. is supported by Leukemia Research Foundation, Gabrielle's Angel Foundation for Cancer Research, the Hairy Cell Research Foundation, and the SASS Foundation for Medical Research. J.I.M.-S. is funded by the Worldwide Cancer Research (grant 16-1285) and the European Hematology Association.
Authorship
Contribution: C.C.O. and J.I.M.-S. wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher C. Oakes, Division of Hematology, Department of Internal Medicine, The Ohio State University, 400 W. 12th Ave, Suite 455, Columbus, OH 43210; e-mail: christopher.oakes@osumc.edu.