Key Points
Hypoxia regulates EHT through Hif-1α and Hif-2α.
The Hif pathway functions upstream of Notch signaling in HSC formation.
Abstract
During development, hematopoietic stem cells (HSCs) derive from specialized endothelial cells (ECs) called hemogenic endothelium (HE) via a process called endothelial-to-hematopoietic transition (EHT). Hypoxia-inducible factor-1α (HIF-1α) has been reported to positively modulate EHT in vivo, but current data indicate the existence of other regulators of this process. Here we show that in zebrafish, Hif-2α also positively modulates HSC formation. Specifically, HSC marker gene expression is strongly decreased in hif-1aa;hif-1ab (hif-1α) and in hif-2aa;hif-2ab (hif-2α) zebrafish mutants and morphants. Moreover, live imaging studies reveal a positive role for hif-1α and hif-2α in regulating HE specification. Knockdown of hif-2α in hif-1α mutants leads to a greater decrease in HSC formation, indicating that hif-1α and hif-2α have partially overlapping roles in EHT. Furthermore, hypoxic conditions, which strongly stimulate HSC formation in wild-type animals, have little effect in the combined absence of Hif-1α and Hif-2α function. In addition, we present evidence for Hif and Notch working in the same pathway upstream of EHT. Both notch1a and notch1b mutants display impaired EHT, which cannot be rescued by hypoxia. However, overexpression of the Notch intracellular domain in ECs is sufficient to rescue the hif-1α and hif-2α morphant EHT phenotype, suggesting that Notch signaling functions downstream of the Hif pathway during HSC formation. Altogether, our data provide genetic evidence that both Hif-1α and Hif-2α regulate EHT upstream of Notch signaling.
Introduction
Hematopoiesis is the process of blood cell production, and it occurs in 2 waves known as primitive and definitive.1 In vertebrate embryos, the definitive wave occurs in several locations, including the ventral wall of the dorsal aorta (VDA),2,3 where a specific subset of endothelial cells (ECs), referred to as hemogenic endothelium (HE), resides. The HE undergoes a process called endothelial-to-hematopoietic transition (EHT) to give rise to hematopoietic stem cells (HSCs). In this process, some ECs acquire hematopoietic potential and subsequently extrude from the VDA to form HSCs.4-6
The transcription factor RUNX1 is one of the main players that positively regulates EHT.6,7 In mice, Runx1 is required for the formation of intra-arterial clusters and consequently of HSCs.8 Accordingly, in zebrafish, both knockdown9,10 and knockout11 of runx1 abrogate definitive hematopoiesis from the VDA. A well-known regulator of Runx1 expression is the Notch signaling pathway. In both mice12 and zebrafish,13 loss-of-function models for Notch1 exhibit a pronounced reduction in Runx1 expression and consequently impaired definitive hematopoiesis. Mice that lack Gata2, a zinc-finger transcription factor gene directly regulated by NOTCH1,14 show dramatic defects in EHT.15 The Notch1-Gata2-Runx1 axis is conserved in zebrafish, because Notch1a and Notch1b have been shown to regulate gata2b expression,13 which in turn positively modulates runx1 transcription in a cell-autonomous manner.16,17 Downstream of RUNX1 in the EHT molecular cascade lies cMyb.18 Loss-of-function murine19-21 and fish22 models for cMyb exhibit defects in several steps of hematopoiesis, including HSC differentiation and maintenance.
In recent years, the hypoxia-inducible factor (Hif) pathway has been suggested to be involved in EHT. In zebrafish, experiments that use morpholinos (MOs) against hif-1ab and treat the embryos with chemicals such as dimethyloxalylglycine (DMOG) to stabilize Hif-α proteins have led to the suggestion that the Hif pathway is a positive regulator of runx1 and cmyb expression.23 These data were later supported by studies in mice in which a conditional knockout of Hif-1α in vascular endothelial cadherin-expressing cells caused a significant but not complete reduction in HSC numbers, suggesting that other players are involved.24
In this study, we generated zebrafish mutants for hif-2α (hif-2aa;hif-2ab), which exhibit a reduction in ECs undergoing EHT, and show that hif-1α and hif-2α both play a role in this process. Exposure to hypoxia strongly induces EHT in wild-type (WT) embryos and partially rescues the phenotype in hif-1α and hif-2α single mutants. In addition, we provide evidence for the hypoxia-Hif pathway acting upstream of Notch signaling in EHT. Altogether, our data provide new insights into the role of the Hif pathway in HSC formation and refine the hierarchy of the EHT transcription cascade.
Methods
Zebrafish handling
Zebrafish were maintained and embryos were obtained and raised under standard conditions25 in accordance with institutional (Max-Planck-Gesellschaft) and national ethical and animal welfare guidelines.
Generation of hif-2aabns231 and hif-2abbns232 mutants
pT7-gRNA and pT3TS-nlsCas9nls vectors were purchased from Addgene.32 A guide RNA (gRNA) was designed to target exon 8 of hif-2aa using the CRISPR design tool (http://crispr.mit.edu/). Generation of gRNA and Cas9 messenger RNA (mRNA) was performed as described previously.33 A transcription activator-like effector nuclease (TALEN) targeting exon 3 of hif-2ab was designed and cloned using Golden Gate assembly into the pCS2TAL3RR or pCS2TAL3DD expression vectors.34 Generation of TALEN arm mRNAs was performed as described previously.33 Mutant alleles were identified by sequencing polymerase chain reaction (PCR) products; bands of 281 and 256 bp for hif2-aa and hif2-ab, respectively, were purified from a 1% agarose gel and sequenced. gRNA sequences, TALEN repeat of variable diresidues, and primers used for genotyping are listed in supplemental Tables 1, 2, and 3, respectively (available on the Blood Web site).
Generation of notch1abns135 mutant and genotyping of notch1bsa11236 mutant
A TALEN targeting exon 11 of notch1a was designed and cloned by using Golden Gate assembly into the pCS2TAL3RRR or pCS2TAL3DDD expression vectors.34 Mutant alleles were identified by high resolution melt analysis35 of PCR products. notch1b mutants were ordered from the European Zebrafish Resource Center. Heterozygous fish were distinguished from WT and homozygous mutant fish by high resolution melt analysis35 of PCR products. Homozygous mutant and WT fish display a similar melting profile, so to distinguish them, a PCR product of 518 bp was purified from 1% agarose gel and was sent for sequencing. TALEN repeat of variable diresidues and primers for genotyping are listed in supplemental Tables 2 and 3, respectively.
Hypoxia chamber treatment
The hypoxia chamber was flushed with nitrogen gas to reach 3% O2 concentration (C-Chamber, ProOX 110, ProCO2 from Biospherix) at 28°C. 50-mm Petri dishes containing 4 mL of embryo water were pre-equilibrated in the chamber overnight. Treatments started at 28 hours post fertilization (hpf) for 8 hours.
Imaging
Images and videos were acquired by using an LSM800 confocal laser scanning microscope (Zeiss) or high-end stereoscopic microscopes (Nikon SMZ25) after embryos were anesthetized with a low dose of tricaine and immobilized in 1% low-melting agarose in glass-bottom Petri dishes (MatTek Corporation, Ashland, MA). Time-lapse images were recorded every 15 minutes for a maximum period of ∼14 hours. The microscope stage was enclosed in a temperature-controlled chamber at 28°C.
Quantitative PCR
Quantitative PCR (qPCR) was performed on complementary DNA obtained from whole embryo or isolated trunk RNA extracted by using TRIZOL (Life Technologies).36 For isolation of embryo trunks, a pool of 36 hpf embryos (∼100 per tube) was subjected to pipetting through a 200-μL pipette for 90 seconds. After centrifugation at 100g for 3 minutes, the solution was transferred into a dish containing cold Hank’s salt solution to select and collect only the trunks under a dissecting microscope. Complementary DNA was synthesized by using SuperScript III RT (Invitrogen) with 300 μg of RNA. A Bio-Rad Real-Time PCR System was used for qPCR experiments. All reactions were performed in 3 technical replicates, and the results represent 3 independent biological samples. qPCR primers are listed in supplemental Table 4.
Whole-mount in situ hybridization
Whole-mount in situ hybridization (WISH) was performed as described previously.37 Primers used to make WISH probes are listed in supplemental Table 5. If genotyping was necessary after imaging, the embryos were placed individually into PCR tubes with 10 μL of 50 mM NaOH. The samples were incubated at 95°C for 20 to 30 minutes (vortexing), and then 1 μL of 1 M tris(hydroxymethyl)aminomethane [pH 8.0] was added to the solution. The extracted DNA was used for genotyping.
MO injections
MOs targeting hif-1aa,38 hif-1ab,39 hif-2aa,38 hif-2ab,38 and the control standard MO were purchased from Gene Tools (Eugene, OR). After optimal titration (no overt toxic effects were observed), hif-1aa and hif-1ab MOs were injected at the 1-cell stage at 0.5 ng per embryo, and hif-2aa and hif-2ab MOs were injected at 1 ng per embryo. MO sequences are listed in supplemental Table 6.
Overexpression of Notch intracellular domain
Progeny from Tg(-1.5hsp70l:GAL4)kca4;(5xUAS-E1b:6xMYC-notch1a)kca3 fish as well as WT embryos were subjected to 37°C heat shock at 14 or 20 hpf for 50 minutes. The embryos were then kept at 28°C until 36 hpf, when they were fixed and processed for WISH. Tg(fli1a:GAL4FF)ubs4 were mated to (5xUAS-E1b:6xMYC-notch1a)kca3, and the resulting embryos were processed for WISH at 36 hpf. The embryos were selected according to their phenotype and were photographed and subsequently genotyped for Tg(fli1a:GAL4FF)ubs4 and Tg(5xUAS-E1b:6xMYC-notch1a)kca3. Primers used for genotyping are listed in supplemental Table 3.
Overexpression of vegfaa and evi1
Full-length vegfaa121 and evi1 coding sequences were cloned into pCS2+. mRNAs were synthesized by using an mMESSAGE mMACHINE SP6 in vitro transcription kit. Zebrafish embryos were injected at the 1-cell stage with 50 pg of vegfaa mRNA or 250 pg of evi1 mRNA.
Quantification and statistical analysis
For the quantification shown in Figure 2D, we counted the number of cells co-expressing Tg(cmyb:EGFP) and Tg(kdrl:mCherry) within as well as emerging from the VDA at 36 hpf in a 10-somite-long trunk area above the yolk extension. hif-1aa+/−;hif-1ab+/− fish were identified by genotyping and incrossed, and the resulting embryos were processed for the WISH experiments shown in Figures 1 and 4. The same procedure was followed for hif-2aa+/−;hif-2ab+/−, and results are shown in Figures 1 and 4. For the experiments shown in Figure 5 and supplemental Figure 14, notch1a+/− fish were incrossed, and the resulting progeny were processed for WISH. Next, the embryos were selected on the basis of their phenotype, photographed, and processed for genotyping as described above. The same procedure was followed for notch1b+/− fish; results are shown in Figure 5 and supplemental Figure 14. The quantification of phenotypes observed by WISH is represented in the figures as n/n (number of embryos showing representative phenotype/total number of embryos examined). Statistical analysis was performed by using GraphPad software. Data in bar graphs represent mean with standard error of the mean or standard deviation. P values were calculated by Student t test for single comparisons of normally distributed data.
hif-1α−/−and hif-2α−/−exhibit decreased runx1 and cmyb expression. (A-L) Brightfield images of WISH for runx1, cmyb, and runx1/cmyb expression in (A-C and G-I) WT sibling, (D-F) hif-1α−/−, and (J-L) hif-2α−/− embryos (stages shown in the figure). The VDA region is outlined and shown at higher magnification in C, F, I, and L (lateral views). Scale bar, 100 μm. (M-O) Quantification of runx1 (A, D, G, and J), cmyb (B, E, H, and K), and runx1/cmyb (C, F, I, and L) WISH results, showing the percentages of embryos with WT and decreased expression in the genotypes listed. n/n, number of embryos showing representative phenotype/total number of embryos examined.
hif-1α−/−and hif-2α−/−exhibit decreased runx1 and cmyb expression. (A-L) Brightfield images of WISH for runx1, cmyb, and runx1/cmyb expression in (A-C and G-I) WT sibling, (D-F) hif-1α−/−, and (J-L) hif-2α−/− embryos (stages shown in the figure). The VDA region is outlined and shown at higher magnification in C, F, I, and L (lateral views). Scale bar, 100 μm. (M-O) Quantification of runx1 (A, D, G, and J), cmyb (B, E, H, and K), and runx1/cmyb (C, F, I, and L) WISH results, showing the percentages of embryos with WT and decreased expression in the genotypes listed. n/n, number of embryos showing representative phenotype/total number of embryos examined.
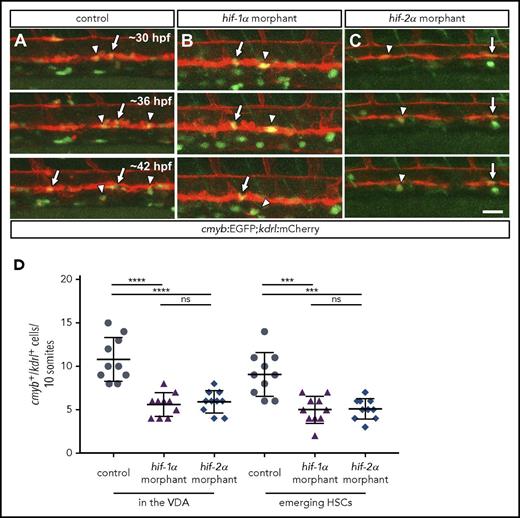
hif-1α and hif-2α morphants show reduced numbers of cmyb:EGFP+;kdrl:mCherry+cells in the VDA. (A-C) Maximal intensity projections of time-lapse confocal images of 36 hpf Tg(cmyb:EGFP);Tg(kdrl:mCherry) control and hif-1α and hif-2α morphants. Arrows point to cmyb:EGFP+;kdrl:mCherry+ cells in the VDA, and arrowheads point to cmyb:EGFP+;kdrl:mCherry+ emerging HSCs. Scale bar, 50 μm. (D) Quantification of cmyb:EGFP+;kdrl:mCherry+ cells in the VDA and emerging HSCs in a 10-somite-long trunk area in control and hif-1α and hif-2α morphants (lateral views) (n = 10 embryos from 3 different clutches). ***P < .001; ****P < .0001. ns, not significant (Student t test).
hif-1α and hif-2α morphants show reduced numbers of cmyb:EGFP+;kdrl:mCherry+cells in the VDA. (A-C) Maximal intensity projections of time-lapse confocal images of 36 hpf Tg(cmyb:EGFP);Tg(kdrl:mCherry) control and hif-1α and hif-2α morphants. Arrows point to cmyb:EGFP+;kdrl:mCherry+ cells in the VDA, and arrowheads point to cmyb:EGFP+;kdrl:mCherry+ emerging HSCs. Scale bar, 50 μm. (D) Quantification of cmyb:EGFP+;kdrl:mCherry+ cells in the VDA and emerging HSCs in a 10-somite-long trunk area in control and hif-1α and hif-2α morphants (lateral views) (n = 10 embryos from 3 different clutches). ***P < .001; ****P < .0001. ns, not significant (Student t test).
Hypoxia is a potent inducer of HSC formation. (A) Schematic representation of the experiment shown in B-H. (B-G) Brightfield images of WISH for (B-C) runx1, (D-E) cmyb, and (F-G) runx1/cmyb expression in 36 hpf WT embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (lateral views). Scale bar, 100 μm. (H) Quantification of runx1 (B-C), cmyb (D-E), and runx1/cmyb (F-G) WISH results, showing the percentages of embryos with WT and increased expression in each condition. (I) qPCR analysis of runx1 and cmyb mRNA levels in 36 hpf WT embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (data for whole embryos is shown; data for dissected trunk is provided in supplemental Figure 11). Values represent mean plus SD (n = 3 biological replicates). Ct values for each gene in WT samples in normoxia: rpl13 = 17.25; runx1 = 28.23; cmyb = 24.48. *P < .05 (Student t test).
Hypoxia is a potent inducer of HSC formation. (A) Schematic representation of the experiment shown in B-H. (B-G) Brightfield images of WISH for (B-C) runx1, (D-E) cmyb, and (F-G) runx1/cmyb expression in 36 hpf WT embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (lateral views). Scale bar, 100 μm. (H) Quantification of runx1 (B-C), cmyb (D-E), and runx1/cmyb (F-G) WISH results, showing the percentages of embryos with WT and increased expression in each condition. (I) qPCR analysis of runx1 and cmyb mRNA levels in 36 hpf WT embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (data for whole embryos is shown; data for dissected trunk is provided in supplemental Figure 11). Values represent mean plus SD (n = 3 biological replicates). Ct values for each gene in WT samples in normoxia: rpl13 = 17.25; runx1 = 28.23; cmyb = 24.48. *P < .05 (Student t test).
Both hif-1α and hif-2α function in EHT. (A) qPCR analysis of runx1 and cmyb mRNA levels in 36 hpf controls, hif-1α−/−, hif-2α−/−, and hif-2α MO-injected hif-1α−/−. Values represent mean plus standard deviation (SD) (n = 3 biological replicates). Ct values for each gene in WT samples: rpl13 = 18.11; runx1 = 27.87; cmyb = 25.81. (B-E) Brightfield images of WISH for runx1/cmyb expression in 36 hpf (B-C) control MO-injected hif-1α−/− and (D-E) hif-2α MO-injected hif-1α−/− in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (lateral views). Scale bar, 100 μm. *P < .05; **P < .01 (Student t test).
Both hif-1α and hif-2α function in EHT. (A) qPCR analysis of runx1 and cmyb mRNA levels in 36 hpf controls, hif-1α−/−, hif-2α−/−, and hif-2α MO-injected hif-1α−/−. Values represent mean plus standard deviation (SD) (n = 3 biological replicates). Ct values for each gene in WT samples: rpl13 = 18.11; runx1 = 27.87; cmyb = 25.81. (B-E) Brightfield images of WISH for runx1/cmyb expression in 36 hpf (B-C) control MO-injected hif-1α−/− and (D-E) hif-2α MO-injected hif-1α−/− in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (lateral views). Scale bar, 100 μm. *P < .05; **P < .01 (Student t test).
Hypoxia does not rescue the EHT defects in notch1a or notch1b mutants. (A) qPCR analysis of notch1a, notch1b, and gata2b mRNA levels in 36 hpf WT embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf. (B) qPCR analysis of notch1a, notch1b, and gata2b mRNA levels in 36 hpf WT, hif-1α−/− and hif-2α−/− embryos. Values represent mean plus SD (n = 3 biological replicates). Ct values for each gene in WT samples: rpl13 = 17.05; notch1a = 21.22; notch1b = 22.21; gata2b = 28.33. (C-J) Brightfield images of WISH for runx1/cmyb expression in 36 hpf (C-D, and G-H) WT siblings, (E-F) notch1a−/− , and (I-J) notch1b−/− embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (lateral views). Scale bar, 100 μm. (K) Quantification of runx1/cmyb WISH results (C-F), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. (L) Quantification of runx1/cmyb WISH results (G-J), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. *P < .05 (Student t test).
Hypoxia does not rescue the EHT defects in notch1a or notch1b mutants. (A) qPCR analysis of notch1a, notch1b, and gata2b mRNA levels in 36 hpf WT embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf. (B) qPCR analysis of notch1a, notch1b, and gata2b mRNA levels in 36 hpf WT, hif-1α−/− and hif-2α−/− embryos. Values represent mean plus SD (n = 3 biological replicates). Ct values for each gene in WT samples: rpl13 = 17.05; notch1a = 21.22; notch1b = 22.21; gata2b = 28.33. (C-J) Brightfield images of WISH for runx1/cmyb expression in 36 hpf (C-D, and G-H) WT siblings, (E-F) notch1a−/− , and (I-J) notch1b−/− embryos in normoxia and after hypoxia exposure for 8 hours starting at 28 hpf (lateral views). Scale bar, 100 μm. (K) Quantification of runx1/cmyb WISH results (C-F), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. (L) Quantification of runx1/cmyb WISH results (G-J), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. *P < .05 (Student t test).
Results
hif-1α and hif-2α mutations lead to a reduced number of HE cells undergoing EHT
To investigate the role of the Hif pathway in EHT, we analyzed hif-1aabns89;hif-1abbns90 and hif-2aabns231;hif-2abbns232 double mutants,33 hereafter abbreviated as hif-1α mutants (hif-1α−/−), and hif-2α mutants (hif-2α−/−), respectively. hif-2aa mutants were generated by using the CRISPR/Cas9 technology40 to target a region in exon 8 that encodes the second PAS domain (supplemental Figure 1A). hif-2ab mutants were generated by using TALEN technology34 to target a region in exon 3 that encodes the first PAS domain (supplemental Figure 2A). We recovered 2 alleles, hif-2aabns231 and hif-2abbns232, carrying out-of-frame mutations that led to a premature stop codon after 26 missense amino acids or 1 missense amino acid, respectively (supplemental Figures 1B-C and 2B-C). To investigate the severity of the mutant alleles, we analyzed hif-2aa and hif-2ab mRNA levels by qPCR. When compared with WT siblings, hif-2aabns231 and hif-2abbns232 mutants display a reduction in the mRNA levels of ∼30% in the mutated gene (supplemental Figures 1D and 2D). To determine whether these mutants exhibit defects in HSC formation, we performed WISH for runx1, one of the earliest known markers of HSC fate. Compared with their WT siblings (Figure 1A,G,M), both hif-1α−/− and hif-2α−/− display a reduction in runx1 expression in the VDA at 30 hpf (Figure 1D,J,M). In addition, we analyzed at 36 hpf the expression levels of cmyb, an important transcription factor gene downstream of Runx1 in the EHT cascade.41 In WT siblings, cmyb was expressed in cells located in both the VDA and caudal hematopoietic tissue (Figure 1B,H,N; supplemental Figure 3A,C,E), and cmyb+ cell numbers were strongly reduced in both hif-1α−/− and hif-2α−/− (Figure 1E,K,N; supplemental Figure 3B,D,E). Consistent with these results, hif-1α−/− and hif-2α−/− embryos (Figure 1F,L,O) exhibited a striking decrease in runx1/cmyb expression levels at 36 hpf compared with WT siblings (Figure 1C,I,O). Similarly, embryos co-injected with hif-1aa and hif-1ab or hif-2aa and hif-2ab MOs (hereafter referred to as hif-1α morphants and hif-2α morphants, respectively) exhibited a decrease in runx1 and cmyb expression levels compared with control MO-injected embryos (supplemental Figure 4). In addition, to test whether the development of the dorsal aorta (DA) was affected in the mutants, we performed WISH for efnb2a (an arterial marker) at 36 hpf. hif-1α−/− and hif-2α−/− exhibited an efnb2a expression level similar to that of their WT siblings (supplemental Figure 5). To further investigate the development of the DA, we analyzed axial vessel formation by confocal imaging using the Tg(kdrl:EGFP)s843 reporter line. We did not observe vascular anomalies in mutant compared with WT embryos (supplemental Figure 6A-C), suggesting that the EHT phenotype observed in hif-1α−/− and hif-2α−/− is not a result of defects in vascular development.
runx1 is also expressed in blood precursors during primitive hematopoiesis.9 To test whether primitive hematopoiesis was affected in hif-1α−/− and hif-2α−/−, we analyzed the expression levels of the lateral plate mesoderm markers runx1 and gata1a several hours before the heart starts beating. We detected WT expression levels of these markers in the lateral plate mesoderm of hif-1α−/− and hif-2α−/− (supplemental Figure 7), indicating that the Hif pathway regulates runx1 expression only during definitive hematopoiesis. To assess whether erythrocyte number or blood flow was affected, we performed WISH for gata1a at 36 hpf as well as brightfield microscopy on live embryos, and we observed WT-like blood development and flow in hif-1α−/− and hif-2α−/− (supplemental Figure 8 and supplemental Videos 1-3).
HSC formation requires 2 sequential steps: HE specification and the emergence of HSCs from this specialized endothelium via EHT.5 To gain further insights into the precise role of hif-1α and hif-2α in this process, we performed time-lapse live imaging on Tg(cmyb:EGFP)zf169;Tg(kdrl:Hsa.HRAS-mCherry)s896 embryos, hereafter Tg(cmyb:EGFP);(kdrl:mCherry), injected with control or hif-1α or hif-2α MOs. By using this transgenic background, it is possible to visualize the HE as cells co-expressing cmyb:EGFP;kdrl:mCherry in the VDA.6 Notably, we observed that hif-1α and hif-2α morphants exhibit a reduced number of cmyb:EGFP;kdrl:mCherry double-positive cells in the VDA compared with control MO-injected embryos (Figure 2A-C; supplemental Videos 4-6), as confirmed by quantification (Figure 2D). Altogether, these observations suggest that Hif-1α and Hif-2α positively regulate runx1 and cmyb expression, thus modulating HE specification, and consequently resulting in a decreased number of emerging HSCs.
Hypoxia strongly induces HSC formation
Mimicking hypoxia with chemicals like DMOG (a pan-hydroxylase inhibitor) reportedly promotes HSC formation in zebrafish.23 To better investigate the impact of hypoxia on EHT, WT embryos were exposed to hypoxia for 8 hours using a hypoxia chamber set at 3% O2, and runx1 and cmyb expression levels were subsequently analyzed by WISH (Figure 3A). WT embryos exposed to hypoxia displayed a strong increase in runx1 and cmyb expression in the VDA at 36 hpf compared with siblings kept in normoxia (Figure 3B-H). To test whether hypoxia exposure affects DA development, we analyzed efnb2a expression by WISH and observed similar expression levels between animals kept in normoxia and those exposed to hypoxia (supplemental Figure 9A-B,E). These data were further confirmed by confocal imaging of vascular development using the Tg(kdrl:EGFP) line (supplemental Figure 6C-D). Moreover, after performing WISH for gata1a at 36 hpf, we did not detect obvious variations in erythrocyte formation after hypoxia exposure (supplemental Figure 10A-B,E), which suggests that the increase in runx1/cmyb expression observed is not a result of an increased number of erythrocytes. To further investigate this phenotype, we performed transcriptional analysis on both whole embryos and isolated trunks at 36 hpf in normoxia and after hypoxia. Our qPCR data show that runx1 and cmyb mRNA levels were significantly upregulated in WT animals after hypoxia compared with siblings in normoxia (Figure 3I; supplemental Figure 11A). Taken together, these data suggest that hypoxia is a potent inducer of EHT through strong induction of runx1 and cmyb expression.
Both hif-1α and hif-2α function in EHT
HIF-2α has been described to have both distinct and overlapping roles with HIF-1α, depending on the biological context.42 hif-1α−/− and hif-2α−/− exhibit similar EHT phenotypes (Figure 1), suggesting a shared function for hif-1α and hif-2α in HE specification in zebrafish. To test this hypothesis, we analyzed runx1 and cmyb mRNA levels in hif-1α−/− and hif-2α−/− by qPCR on 36 hpf whole embryos and isolated trunks and found that they were reduced by ∼30% in both hif-1α−/− and hif-2α−/− compared with WT sibling embryos (Figure 4A; supplemental Figure 11B). Next, we performed hif-2α MO knockdown in hif-1α−/− and assessed runx1 and cmyb mRNA levels at 36 hpf. hif-2α MO-injected hif-1α−/− exhibited a decrease in runx1 and cmyb expression of ∼60% compared with control MO-injected embryos and of ∼50% compared with hif-1α−/− (Figure 4A). These results suggest that hif-1α and hif-2α have similar roles in EHT. To further investigate this hypothesis, we exposed control MO-injected hif-1α−/− embryos to hypoxic conditions and examined runx1/cmyb expression by WISH. Interestingly, after hypoxia exposure, runx1/cmyb expression levels were increased compared with normoxia (Figure 4B-C), but this expression was not comparable to the strong runx1/cmyb induction observed in WT animals after hypoxia (Figure 3G). This result suggests that hypoxia can rescue, at least in part, the EHT phenotype observed in hif-1α−/−, possibly through hif-2α. To test this hypothesis, we analyzed runx1/cmyb expression levels in hif-2α MO-injected hif-1α−/− in both normoxic and hypoxic conditions. In normoxia, these embryos showed a further decrease in runx1/cmyb expression (Figure 4D) compared with control MO-injected hif-1α−/− (Figure 4B). Importantly, when hif-2α MO-injected hif-1α−/− embryos were exposed to hypoxia, the rescue in runx1/cmyb expression was completely abrogated (Figure 4C,E). These observations show that hypoxia, signaling through Hif-2α, can partially rescue the EHT phenotype observed in hif-1α−/−.
Hypoxia does not rescue the EHT defects in notch1a or notch1b mutants
It has been shown that Notch signaling is a key regulator of EHT. Notch1a and Notch1b modulate gata2b expression, and Gata2b in turn directly regulates runx1 transcription.12-15,43 First, we analyzed whether notch1a, notch1b, and gata2b mRNA levels were influenced by changes in O2 concentration and found that they were strongly induced after hypoxic conditions compared with sibling embryos in normoxic conditions (Figure 5A; supplemental Figure 12). We also tested whether notch1a, notch1b, and gata2b mRNA levels were altered in the absence of hif-1α and hif-2α function. In both hif-1α−/− and hif-2α−/−, we observed a significant reduction in mRNA levels for all 3 genes compared with WT siblings (Figure 5B). These results suggest that Notch acts downstream of the Hif pathway during EHT in zebrafish. To test this hypothesis, we took advantage of notch1a and notch1b mutant alleles available in our laboratory. notch1a−/− was generated by TALEN technology targeting a region in exon 11 that encodes an EGF-like repeat in the extracellular portion of the receptor (supplemental Figure 13A). We recovered an allele, notch1abns135, carrying an out-of-frame mutation that led to a premature stop codon after 19 missense amino acids (supplemental Figure 13B-C). notch1bsa11236 mutants44 contain a T→A transversion in exon 21 (supplemental Figure 13D), which led to a premature stop codon in the extracellular part of the protein (supplemental Figure 13E). First, we tested whether notch1a−/− and notch1b−/− recapitulate the previously published EHT phenotypes using notch1a and notch1b MOs.13 When we incrossed notch1a+/− or notch1b+/−, we observed a reduction in runx1/cmyb and gata2b expression by WISH in both notch1a−/− and notch1b−/− compared with WT siblings (supplemental Figure 14), as reported in notch1a and notch1b morphants.13 We then used these mutants to investigate the molecular hierarchy between Hif and Notch signaling during EHT. We exposed embryos from notch1a+/− or notch1b+/− incrosses to normoxia or hypoxia and analyzed runx1/cmyb expression levels by WISH. WT sibling embryos exposed to hypoxia exhibited an increase in runx1/cmyb expression (Figure 5C-D,G-H,K-L) but notch1a−/− or notch1b−/− embryos did not (Figure 5E-F,I-L). These data show that hypoxia does not induce runx1 or cmyb expression in notch1a−/− or notch1b−/−, unlike what is observed in WT embryos, and thus does not rescue their EHT phenotype. Altogether, our results suggest that Hif and Notch function in the same pathway during EHT.
Activation of Notch signaling in ECs can rescue hif-1α and hif-2α EHT defects
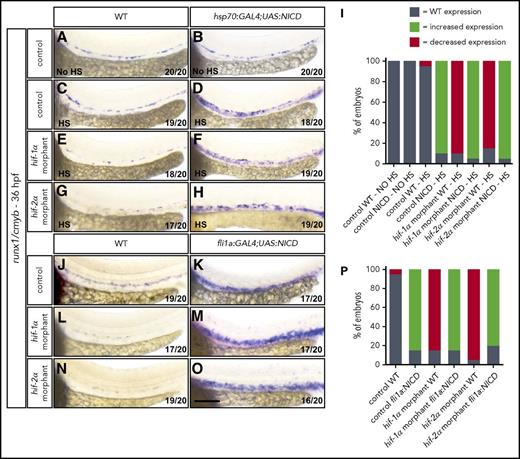
To further investigate the hierarchy between Hif and Notch signaling, we overexpressed the notch1a intracellular domain (NICD) to promote Notch signaling activation in a temporally controlled manner. We used the Tg(-1.5hsp70l:GAL4)kca4;(5xUAS-E1b:6xMYC-notch1a)kca3 line, hereafter, Tg(hsp70:GAL4;UAS:NICD), to drive NICD overexpression upon heat shock treatment. Specifically, we analyzed control MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) animals kept at 28°C and observed that runx1/cmyb expression was indistinguishable between these two populations (Figure 6A-B,I). In parallel, control MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos were subjected to heat shock at 37°C for 50 minutes at both 1445 and 20 hpf46 and were analyzed by WISH for runx1/cmyb at 36 hpf. After heat shock, we did not observe changes in runx1/cmyb expression in control WT embryos (Figure 6C), but Tg(hsp70:GAL4;UAS:NICD) embryos exhibited enhanced expression of HSC markers in the VDA (Figure 6D,I; supplemental Figure 15A-B,G). To assess DA formation, we first examined efnb2a expression by WISH and found that it was slightly increased upon heat shock; however, it was not expressed ectopically compared with Tg(hsp70:GAL4;UAS:NICD) embryos left at 28°C (supplemental Figure 9C-E). To investigate the vascular pattern, we analyzed Tg(kdrl:EGFP) expression in Tg(hsp70:GAL4;UAS:NICD) embryos and did not detect vascular abnormalities after heat shock (supplemental Figure 6E-F). Furthermore, to analyze whether erythrocyte formation was affected, we performed WISH for gata1a at 36 hpf and observed no significant changes in Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock (supplemental Figure 10C-E). Next, we analyzed runx1/cmyb levels in hif-1α or hif-2α MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos. In nontransgenic hif-1α and hif-2α morphants, we observed a decrease in runx1/cmyb expression (supplemental Figure 4), which was not influenced by heat shock treatment (Figure 6E,G,I). Conversely, in Tg(hsp70:GAL4;UAS:NICD) embryos injected with hif-1α or hif-2α MOs, we could detect a strong induction of runx1/cmyb expression after heat shock (Figure 6F,H,I; supplemental Figure 15C-G), thereby rescuing the defects observed in hif-1α and hif-2α morphants. To further investigate this ability of Notch signaling to rescue, we took advantage of the double transgenic line Tg(fli1a:GAL4FF)ubs4;(5xUAS-E1b:6xMYC-notch1a)kca3, hereafter, Tg(fli1a:GAL4;UAS:NICD), to overexpress NICD specifically in ECs. We observed high expression levels of runx1/cmyb in control MO-injected Tg(fli1a:GAL4;UAS:NICD) embryos compared with control MO-injected WT siblings (Figure 6J-K,P). Next, we injected hif-1α or hif-2α MOs into WT and Tg(fli1a:GAL4;UAS:NICD) embryos. In nontransgenic hif-1α and hif-2α morphants, we observed a reduction in runx1/cmyb expression (Figure 6L,N,P). Conversely, in Tg(fli1a:GAL4;UAS:NICD) embryos injected with hif-1α or hif-2α MOs, the expression of runx1/cmyb was strongly induced, rescuing the hif-1α and hif-2α morphant phenotypes (Figure 6M,O-P). These data indicate that global as well as endothelial-specific NICD overexpression can rescue the EHT defects observed in Hif-1α and Hif-2α loss-of-function models.
NICD overexpression in ECs rescues the EHT defects in hif-1α and hif-2α morphants. (A-H) Brightfield images of WISH for runx1/cmyb expression in (A-B) 36 hpf control MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos at 28°C, (C-D) control MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock, (E-F) hif-1α MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock, and (G-H) hif-2α MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock. (I) Quantification of runx1/cmyb WISH results (A-H), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. (J-O) Brightfield images of WISH for runx1/cmyb expression in (J-K) 36 hpf control MO-injected WT and Tg(fli1a:GAL4;UAS:NICD) embryos, (L-M) hif-1α MO-injected WT and Tg(fli1a:GAL4;UAS:NICD) embryos, and (N-O) hif-2α MO-injected WT and Tg(fli1a:GAL4;UAS:NICD) embryos (lateral views). (P) Quantification of runx1/cmyb WISH results (J-O), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. Scale bar, 100 μm.
NICD overexpression in ECs rescues the EHT defects in hif-1α and hif-2α morphants. (A-H) Brightfield images of WISH for runx1/cmyb expression in (A-B) 36 hpf control MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos at 28°C, (C-D) control MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock, (E-F) hif-1α MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock, and (G-H) hif-2α MO-injected WT and Tg(hsp70:GAL4;UAS:NICD) embryos after heat shock. (I) Quantification of runx1/cmyb WISH results (A-H), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. (J-O) Brightfield images of WISH for runx1/cmyb expression in (J-K) 36 hpf control MO-injected WT and Tg(fli1a:GAL4;UAS:NICD) embryos, (L-M) hif-1α MO-injected WT and Tg(fli1a:GAL4;UAS:NICD) embryos, and (N-O) hif-2α MO-injected WT and Tg(fli1a:GAL4;UAS:NICD) embryos (lateral views). (P) Quantification of runx1/cmyb WISH results (J-O), showing the percentages of embryos with WT, increased expression, and decreased expression in each condition. Scale bar, 100 μm.
To further characterize the link between Hif and Notch signaling, we tested whether the overexpression of evi1 or vegfaa was able to rescue the EHT phenotype observed in hif-1α and hif-2α mutants. As previously described,46 the overexpression of these 2 genes leads to an increase in notch1b expression levels in HE during EHT. Interestingly, the injection of evi1 or vegfaa mRNA in hif-1α−/− and hif-2α−/− rescued runx1/cmyb expression (supplemental Figure 16), which suggests that evi1 and vegfaa function downstream or in parallel to the Hif pathway during EHT. Altogether, our observations suggest that Notch signaling functions downstream of hypoxia and the Hif pathway during EHT.
Discussion
The differentiation of cell lineages from pluripotent stem cells has been achieved in vitro by inducing the expression of transcription factors or exposing pluripotent cells to morphogens. Notably, most of the molecules identified to promote cell specification in vitro were reported to play a role during embryonic development. Many groups have been trying to develop HSC-like cells in vitro but have not yet obtained successful long-term engraftments in mouse recipients.47-52 One recent publication showed advances in the robustness of HSC engraftment into mouse hosts, leaving room for improvement.53 Another recent article described a method to reprogram adult ECs to HSCs through transient expression of transcription factors and vascular niche-derived angiocrine signals.54 A better understanding of all the players involved in hematopoiesis and how intrinsic and extrinsic factors are coordinated in the vascular niche could have a significant impact on the generation of therapy-grade HSCs. Here, by using a genetic approach in zebrafish, we describe a novel role for hif-2α in HSC formation and confirm previous reports on hif-1α in this process. Moreover, we provide evidence for the Hif pathway functioning upstream of Notch signaling during EHT (Figure 7).
Proposed model. Schematic illustration of the role of the Hif pathway during EHT. We propose that Hif functions upstream of Notch1 signaling in EHT.
Proposed model. Schematic illustration of the role of the Hif pathway during EHT. We propose that Hif functions upstream of Notch1 signaling in EHT.
In a previous article, Harris et al23 showed that the stabilization of Hif-α subunits with chemicals such as DMOG can strongly induce runx1 and cmyb expression during EHT. These experiments suggested a role for the Hif pathway in this process but did not explore the role of O2 concentration and could not exclude possible adverse effects of DMOG.55 In our study, we exposed zebrafish embryos to hypoxic conditions using a hypoxia chamber at 3% O2 concentration. Embryos exposed to hypoxia exhibited increased expression of HSC markers in the VDA, suggesting that hypoxia is a potent inducer of EHT in zebrafish. These data are consistent with a recent report showing that aortic ECs emerging from the DA in mice are hypoxic.24
The Hif-1α pathway has been suggested to regulate EHT on the basis of MO studies.23,24 Recently, several publications have highlighted the phenotypic differences between morphants and mutants56-58 and the importance of validating the morphant phenotypes described in the past.59 Here, using hif-1α and hif-2α mutants, we confirm the previous MO studies on the role of hif-1α during EHT and describe a novel role for hif-2α in this process. Importantly, by using time-lapse imaging, we found that the reduction in HE cell number in the VDA is the likely cause of the HSC defects in hif-1α−/− and hif-2α−/−, a novel observation. HIF-1α and HIF-2α have been described to have both distinct and overlapping roles, depending on the biological context.42 A previous article has reported a partial loss of HSC number in Hif-1α-mutant mice.24 Here, we suggest that this observation may be due, at least in part, to HIF-2α, which may be able to compensate partially for the loss of HIF-1α.
HIF is mainly involved in the transcriptional activation of hypoxia-responsive genes, and recent studies have suggested that the HIF transcriptional complex may interact with certain signaling pathways, including Notch. Here, our data suggest that Hif and Notch function in the same pathway during EHT. Specifically, NICD overexpression in ECs could rescue the HSC defects observed in hif-1α and hif-2α mutants, suggesting that Notch functions downstream of Hif. Given the multiple roles of Notch signaling in developmental processes, it is possible that Notch could also be acting upstream or in parallel to Hif. The relationship between the Notch and Hif pathways is also context specific and has been explored in different settings; a recent article showed that in crystal cells (a blood cell type in Drosophila), sima, the ortholog of Hif-1α, activates Notch receptor in both normoxic and hypoxic cells in a cell-autonomous manner.60
Our study provides new insights into the role of hypoxia and the Hif pathway in definitive hematopoiesis, and our genetic analyses provide a link between the Hif pathway and Notch signaling. These data help further understand the players and microenvironment that modulate HSC formation, which will have an impact on the ways HE-derived HSCs are generated for therapeutic purposes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michelle M. Collins, Anabela Bensimon-Brito, Jason Kuan Han Lai, Felix Gunawan, Hyouk-Bum Kwon, S. Javad Rasouli, Kenny Mattonet, Silvia Parajes Castro, and Ryota Matsuoka for sharing reagents and protocols; Francesca Luzzani for support in some experiments; members of the Stainier Laboratory, in particular Rubén Marín-Juez, for helpful discussions.
The Max Planck Society, International Max Planck Research Schools, Deutsche Forschungsgemeinschaft (SFB 834) provided funding.
Authorship
Contribution: C.G., M.M., and D.Y.R.S. designed experiments and analyzed data; C.G. and M.M. conducted experiments; A.R. contributed an unpublished reagent; and C.G., M.M., and D.Y.R.S. wrote the paper with feedback from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Didier Y.R. Stainier, Max Planck Institute for Heart and Lung Research, Developmental Genetics (Dept. III), Ludwigstraße 43, 61231 Bad Nauheim, Germany; e-mail: didier.stainier@mpi-bn.mpg.de.
References
Author notes
C.G. and M.M. contributed equally to this study.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal