Key Points
Nbeal2 interacts with Dock7, Sec16a, and Vac14; and missense variants that cause GPS disrupt the binding of Dock7 and Vac14.
The level of the α-granule protein Dock7 in platelets from Nbeal2−/− mice and GPS cases is reduced and its signaling pathway is dysregulated.
Abstract
Mutations in NBEAL2, the gene encoding the scaffolding protein Nbeal2, are causal of gray platelet syndrome (GPS), a rare recessive bleeding disorder characterized by platelets lacking α-granules and progressive marrow fibrosis. We present here the interactome of Nbeal2 with additional validation by reverse immunoprecipitation of Dock7, Sec16a, and Vac14 as interactors of Nbeal2. We show that GPS-causing mutations in its BEACH domain have profound and possible effects on the interaction with Dock7 and Vac14, respectively. Proximity ligation assays show that these 2 proteins are physically proximal to Nbeal2 in human megakaryocytes. In addition, we demonstrate that Nbeal2 is primarily localized in the cytoplasm and Dock7 on the membrane of or in α-granules. Interestingly, platelets from GPS cases and Nbeal2−/− mice are almost devoid of Dock7, resulting in a profound dysregulation of its signaling pathway, leading to defective actin polymerization, platelet activation, and shape change. This study shows for the first time proteins interacting with Nbeal2 and points to the dysregulation of the canonical signaling pathway of Dock7 as a possible cause of the aberrant formation of platelets in GPS cases and Nbeal2-deficient mice.
Introduction
Cellular secretion is a key biological event involved in many physiological processes, including hemostasis.1,2 Secretory cells store granules, which are released according to predetermined temporal and spatial programs.3 The function of secretory cells is to a large extent linked to their types of granules, their content, and the signaling events leading to secretion. Therefore, the ontology and homeostasis of granules must be highly regulated (ie, from the formation of the vesicles themselves and cargo sorting through cytoplasmic maintenance of formed vesicles and eventual exocytosis).4,5
Blood platelets are anucleated secretory cells originating from megakaryocytes (MKs).6 Their primary role is in hemostasis,7 but they are also relevant in other pathophysiological pathways.8-12 Platelet function is mediated by the release of their 3 types of granules upon vascular injury.13 Aberrations in any of these 3 granule types lead to inability of platelets to sustain hemostasis, resulting in bleeding.14 Defects in lysosomes are observed in Chediak-Higashi syndrome and caused by mutations in LYST encoding a member of the family of Beige and Chediak-Higashi (BEACH)-domain (BEACH hereafter) containing proteins.15 Aberrations of platelet δ-granules are observed in Hermansky-Pudlak syndrome caused by mutations in 11 genes encoding proteins with roles in the formation and trafficking of granules.16 Haploinsufficiency of another BEACH family member, Neurobeachin (NBEA), is associated with severe autism and atypical δ-granules in humans17 and Nbea heterozygous mice.18
The α-granules are the most abundant organelles and granules in platelets. They contain hundreds of proteins with important functions in hemostasis and wound healing.8-12,19 Loss of function variants of the BEACH-domain gene NBEAL2 in gray platelet syndrome (GPS) cases leads to absence of platelet α-granules.20-23 Nbeal2−/− mice present with a faithful copy of human GPS with the typical large platelets of grayish appearance, and interestingly, neutrophils and mast cells are also dysfunctional.24,25 In vivo studies highlight the role of Nbeal2 beyond hemostasis, particularly in inflammation, innate immunity, and cancer.24,26-28 However, the molecular function of Nbeal2 protein has not been explored.
The BEACH proteins Lyst, Nbea, and Nbeal2 are deemed essential for molecular processes underlying the formation, retention, and secretion of the 3 types of granules in platelets.29 All 9 BEACH genes encode large proteins between 2000 and 4500 residues containing the Armadillo-type, Concanavalin-A lectin, Pleckstrin Homology (PH), and WD40 repeat domains in addition to the typical BEACH domain.29 This domain of ∼35 kDa is unique to the BEACH family, is highly conserved in eukaryotes, and does not share sequence homology with other proteins.15 It is characterized by a unique hydrophobic secondary structure that cannot be classified as either β-strands or random coils30 and strongly interacts with its neighboring PH domain, but the significance of this interaction remains elusive.31
Here, we use expression of Nbeal2 domains in human cells to define its proximal interactome with confirmation of Dock7, Sec16a, and Vac14 as Nbeal2 binding partners by different biochemical and cellular approaches. We show that GPS-causing mutations strongly reduce the interaction of Dock7 with the BEACH domain, and platelets from Nbeal2−/− mice and GPS patients are nearly devoid of Dock7 with a direct impact on platelet function.
Materials and methods
Short descriptions of the methods used are provided in this section, and complete details are given in the supplemental Methods, available on the Blood Web site.
Cell culture and stable clones
Human embryonic kidney 293T cells (HEKs) and CHRF-288 cells (CHRFs) were cultured following AATC procedures. Stable transfectants ectopically expressing the protein domains under study were generated by infection with lentiviral particles. HEKs were transduced at a multiplicity of infection of 10 transducing units per cell in the presence of 1 μg/mL polybrene. CHRFs were transduced at a multiplicity of infection of 50 transducing units per cell in the presence of 1 μg/mL protamine sulfate.
Tandem affinity purification
Cells were collected and washed twice with cold phosphate-buffered saline (PBS), and pellets were snap-frozen on dry ice and kept at −80°C until immunoprecipitation. Pellets were resuspended with lysis buffer containing 0.1% NP-40 and protease inhibitor cocktail. Tandem affinity purification and sample preparation for mass spectrometry analysis were performed as previously described.32
Mass spectrometry and data analysis
Peptides were redissolved in 0.5% formic acid and analyzed with on-line nano liquid chromatography tandem mass spectrometry on an LTQ FT mass spectrometer (Thermo-Fisher Scientific) coupled with an Ultimate 3000 Nano/Capillary LC System (Dionex). Proteins frequently identified from tandem affinity purifications of unrelated targets in embryonic stem cells were discarded.
The mass spectrometry proteomics data can be retrieved from the ProteomeXchange Consortium via the PRIDE33 partner repository with the dataset identifier PXD006091.
Immunoprecipitation of endogenous proteins
Total cell lysates were generated as above, and proteins of interest were immunoprecipitated using Protein G Dynabeads (Invitrogen) and selected antibodies.
Proximity ligation assays
Human MKs were generated from CD34+ hematopoietic stem cells as stated in supplemental Methods. Proximity ligation assays (PLAs) were performed following the manufacturer’s protocol.
Mice
Adult animals were used, including age-matched controls of the same genetic background. This study has been regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (Home Office PPL70/8406). A minimum of 5 different mice per experiment was used unless otherwise stated.
Isolation of platelets
Mouse blood was withdrawn from the inferior vena cava in either EDTA (5 mM final) or ACD (acid-citrate-dextrose; 111 mM glucose, 71 mM citric acid, 116 mM sodium citrate), whereas blood from GPS patients and controls was taken as previously reported.21 In both cases, platelets were washed using Tyrode’s buffer pH 6.5.
Immunoblots
HEKs, CHRFs, and platelets were lysed by addition of FLAG tandem affinity purification (FTAP)-lysis buffer (0.1% NP-40, 150 mM NaCl, 10 mM Tris, pH 8) containing protease inhibitor cocktail and incubated on ice for 10 minutes before centrifugation at 13 000 rpm for 15 minutes at 4°C.
Analysis of active Cdc42 and Rac1
Platelets were washed as above and adjusted to 3 × 105 platelets per microliter in Tyrode’s buffer pH 7.35. Stimulations with vehicle (PBS) and thrombin (T4648; Sigma, UK) were done at room temperature for different time points, and lysis buffer provided by assay kit (BK127 and BK128, Cytoskeleton) was added. Subsequent steps were carried out following the manufacturer’s protocol. Absorbance at 490 nm was read using an MP5 plate reader (Molecular Devices, UK).
F-actin formation in platelets
Washed platelets were activated for 10 minutes and then fixed, permeabilized, incubated with 488-Phalloidin for 20 minutes, and read in a Gallios flow cytometer (Beckman Coulter, UK).
Platelet spreading on fibrinogen
A solution of 100 μg/mL of fibrinogen (FIB3; Enzyme Research, UK) in PBS was used to coat coverslips (12392128; Thermo-Fisher Scientific) overnight at 4°C. Platelets at 5 × 103/μL in Tyrode’s buffer pH 7.35 were seeded onto the coverslips, and platelet agonists or vehicle was added.
Transmission electron microscopy and gold-labeling
Confocal microscopy
Cells grown on coverslips were fixed with 2% paraformaldehyde-PBS for 30 minutes at room temperature and washed twice with PBS.
Statistics
Results are shown as mean ± standard error of the mean (SEM). Statistical analysis was performed using the unpaired Student t test. P values < .05 were considered statistically significant.
A complete list of primary antibodies used in this study is provided in supplemental Table 1.
Results
The Nbeal2 interactome
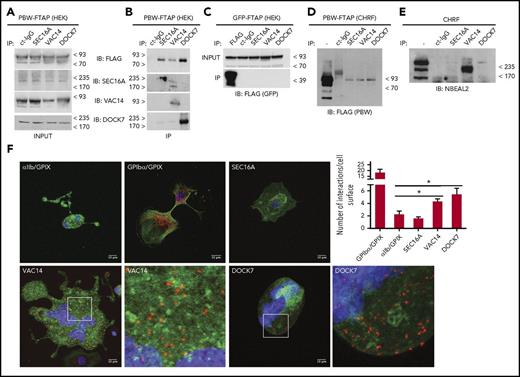
To identify the putative interacting proteins of Nbeal2 (the interactome hereafter), a construct encompassing the PH, BEACH, and WD40 (PBW) domains and a FTAP tag32 (Figure 1A) was used to transform HEKs (Figure 1B), which endogenously express Nbeal2 (supplemental Figure 1A). An FTAP-tagged green fluorescent protein (GFP) construct was used as a control (Figure 1A-B). Cells were then used in a 2-step (FLAG and calmodulin binding peptide) sequential affinity purification, and the resulting eluate was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by mass spectrometry analysis (Figure 1C). Proteins identified in GFP samples and from an “in-house contaminant” database as stated in the “Materials and methods” were removed from further analysis. The classification of the remaining 132 proteins (129 entries by gene name excluding NBEAL2; supplemental Table 2) according to the most frequent gene ontology terms shows an array of biological functions (Figure 1D).
The Nbeal2 interactome. (A) Diagram showing the functional domains of Nbeal2 (ARM, Armadillo; ConA, Concanavalin A; WD40 repeat [WD40]). Construct 1 (PBW-FTAP) encompasses the PH, BEACH, and aminoterminal WD40 domains and the FTAP tag. Construct 2 (GFP-FTAP) encompasses GFP and FTAP. (B) Ectopic expression of GFP-FTAP and PBW-FTAP in HEKs. (C) FTAP-purification of GFP-FTAP and PBW-FTAP from HEKs. Purified proteins were resolved by SDS-PAGE, visualized by Coomassie blue stain, and analyzed by liquid chromatography-mass spectrometry. Some lanes in gels from panels B and C were deleted and replaced by black vertical lines. (D) Identified proteins (n = 129) were classified according to their protein class using gene ontology terms. (E) PPIN; 637 bait proteins representing the processes of hemostasis, megakaryopoiesis, and platelet formation were used to retrieve 5112 nodes (proteins) and 26 702 edges (biochemical reactions) by Reactome and IntAct. Orange nodes (n = 91) are interactors present in the PPIN, and the purple nodes (n = 38) have interactions with 1 or more proteins of the PPIN. Nbeal2 is shown in black and Dock7, Sec16a, and Vac14 are shown as orange triangles. (F-H) Subnetworks extracted from the PPIN in panel E for Dock7 (F), Sec16a (G), and Vac14 (H). The expression levels for the corresponding genes determined by the sequencing of RNA of human MKs and expressed as log2 FPKM (fragments per kilobase of transcript per million mapped reads) is presented in white-to-green color, and only nodes corresponding to genes with FPKM > 1 in MKs are depicted (see color bar for relative FPKM values).43 The gray/black (E-H), light blue (F-H), and dark blue (F-H) edges are based on the results of pull-down experiments reported in this study, Reactome and IntAct, respectively. A Cytoscape file to generate an interactive version of the PPIN, containing gene expression levels and other annotation features, is available from the supplemental data. MW, molecular weight.
The Nbeal2 interactome. (A) Diagram showing the functional domains of Nbeal2 (ARM, Armadillo; ConA, Concanavalin A; WD40 repeat [WD40]). Construct 1 (PBW-FTAP) encompasses the PH, BEACH, and aminoterminal WD40 domains and the FTAP tag. Construct 2 (GFP-FTAP) encompasses GFP and FTAP. (B) Ectopic expression of GFP-FTAP and PBW-FTAP in HEKs. (C) FTAP-purification of GFP-FTAP and PBW-FTAP from HEKs. Purified proteins were resolved by SDS-PAGE, visualized by Coomassie blue stain, and analyzed by liquid chromatography-mass spectrometry. Some lanes in gels from panels B and C were deleted and replaced by black vertical lines. (D) Identified proteins (n = 129) were classified according to their protein class using gene ontology terms. (E) PPIN; 637 bait proteins representing the processes of hemostasis, megakaryopoiesis, and platelet formation were used to retrieve 5112 nodes (proteins) and 26 702 edges (biochemical reactions) by Reactome and IntAct. Orange nodes (n = 91) are interactors present in the PPIN, and the purple nodes (n = 38) have interactions with 1 or more proteins of the PPIN. Nbeal2 is shown in black and Dock7, Sec16a, and Vac14 are shown as orange triangles. (F-H) Subnetworks extracted from the PPIN in panel E for Dock7 (F), Sec16a (G), and Vac14 (H). The expression levels for the corresponding genes determined by the sequencing of RNA of human MKs and expressed as log2 FPKM (fragments per kilobase of transcript per million mapped reads) is presented in white-to-green color, and only nodes corresponding to genes with FPKM > 1 in MKs are depicted (see color bar for relative FPKM values).43 The gray/black (E-H), light blue (F-H), and dark blue (F-H) edges are based on the results of pull-down experiments reported in this study, Reactome and IntAct, respectively. A Cytoscape file to generate an interactive version of the PPIN, containing gene expression levels and other annotation features, is available from the supplemental data. MW, molecular weight.
To better comprehend its functional role, we used the interaction data to embed Nbeal2 in a protein-protein interaction network (PPIN) representing the processes of hemostasis, megakaryopoiesis, and platelet formation.35 The PPIN of 5112 nodes (proteins) and 27 441 edges (biochemical interactions) was obtained by retrieving first-order interactions from Reactome36 and IntAct37 using a set of 637 bait proteins (baits hereafter). The set of genes encoding the baits resulted from an integrative analysis of a genome-wide association study for platelet traits in 173 480 individuals38,39 and a promoter capture Hi-C assay in human MKs40 ; the 79 genes implicated in Mendelian disorders of hemostasis were also included.14,41 Of the 129 nodes in the Nbeal2 interactome, 91 are present in the PPIN (orange nodes in Figure 1E) and the remaining 38 are first-order interactors with 1 or several proteins of the PPIN (purple nodes in Figure 1E).
Previous studies on proteins from the BEACH family suggest that they may function as scaffolds with a critical role of Nbeal2 in homeostasis of α-granules.26,27,29 Based on this and on a detailed inspection of the PPIN, 10 candidate proteins were selected for validation studies by reverse immunoprecipitation (supplemental Table 2), and the interactions of 3 proteins were confirmed, namely Dock7 (a guanidine exchange factor [GEF]); Sec16a (an endoplasmic reticulum membrane protein); and Vac14 (a regulator of phosphatidyl inositol 3,5-bisphosphate) (triangular orange nodes in Figure 1E). Their first-order interactors are illustrated in the subnetworks in Figure 1F-H (supplemental Figures 2-4 for fully labeled versions).
Dock7, Sec16a, and Vac14 interact with Nbeal2
To confirm the interactions with Nbeal2, we first performed immunoprecipitation of the selected candidates in PBW-FTAP HEKs, and positive results for Dock7, Sec16a, and Vac14 were obtained (Figure 2A- B). These experiments showed that endogenous Sec16a and Vac14 pulled down PBW-FTAP and Dock7, and endogenous Dock7 pulled down PBW-FTAP and Vac14 (Figure 2B; supplemental Figure 5). Assays in GFP-FTAP HEKs, as expected, showed no interactions with any of the 3 proteins (Figure 2C). Second, we used CHRFs, which have a transcriptome reminiscent of MKs42 and a robust Nbeal2 protein expression (supplemental Figure 1B). Lentiviral transduction with PBW-FTAP followed by the same experimental approach as used for HEKs confirmed the interaction with Dock7, Sec16a, and Vac14 in PBW-FTAP CHRFs (Figure 2D). Furthermore, endogenous Nbeal2 was also detected when the 3 interactors were pulled down in nontransduced CHRFs, although the recovery levels for Dock7 and Sec16a were low (Figure 2E). Finally, after the initial successful validation experiments with cell lines, we used PLA in MKs derived from human hematopoietic stem cells.43 The 2 most abundant platelet membrane receptor complexes for fibrinogen (αIIbβ3 integrin) and von Willebrand factor (GPIbα, GPIbβ, GPV, GPIX), respectively, were used to validate the assay. As expected, targeting αIIb and GPIX simultaneously produced a weak PLA signal similar to that using isotype controls, while targeting GPIbα and GPIX generated a strong signal (Figure 2F). When compared with the negative control, we observed a significant increase in PLA signals in Dock7-Nbeal2 and Vac14-Nbeal2 labeled-MKs, further supporting the interactions between these proteins (Figure 2F).
Dock7, Sec16a, and Vac14 interact with Nbeal2. (A-B) Immunoprecipitation (IP) of Dock7, Sec16a, and Vac14 from PBW-FTAP HEKs with total cell lysate (input) in panel A, IP results in panel B. PBW-FTAP was detected by anti-FLAG, and a rabbit isotype immunoglobulin G (IgG) was used as control. Immunoblot with specific antibodies against Dock7, Sec16a, and Vac14 (top blot in panel B). The other 3 blots in panel B show the specific binding of each of the cognate antibodies, and the results with the isotype control immunoglobulin (ct-IgG) are in the first lane of the 4 blots in panel B. (C-E) Similar IP assays as in panel A carried out with HEKs (C) or CHRFs (D-E). (F) PLA with human MKs showing the results for (1) αIIb and GPIX as negative control (top left), (2) GPIbα and GPIX as positive control (top middle), (3) Sec16a (top right); Vac14 (2 bottom left); and Dock7 (2 bottom right); each interactor was tested in combination with Nbeal2. Interactions between proteins are presented as orange dots, and cells were counterstained with Phalloidin (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). For better illustration of the PLA signals, a ×20 magnification of the insets in Vac14 and Dock7 are also shown. (4) Quantitative analysis of the number of interactions per cell surface based on analysis of the PLA signals by Image Studio are presented in the bar graph. Comparisons were made against the negative control. Bars represent mean ± SEM. *P value < .05.
Dock7, Sec16a, and Vac14 interact with Nbeal2. (A-B) Immunoprecipitation (IP) of Dock7, Sec16a, and Vac14 from PBW-FTAP HEKs with total cell lysate (input) in panel A, IP results in panel B. PBW-FTAP was detected by anti-FLAG, and a rabbit isotype immunoglobulin G (IgG) was used as control. Immunoblot with specific antibodies against Dock7, Sec16a, and Vac14 (top blot in panel B). The other 3 blots in panel B show the specific binding of each of the cognate antibodies, and the results with the isotype control immunoglobulin (ct-IgG) are in the first lane of the 4 blots in panel B. (C-E) Similar IP assays as in panel A carried out with HEKs (C) or CHRFs (D-E). (F) PLA with human MKs showing the results for (1) αIIb and GPIX as negative control (top left), (2) GPIbα and GPIX as positive control (top middle), (3) Sec16a (top right); Vac14 (2 bottom left); and Dock7 (2 bottom right); each interactor was tested in combination with Nbeal2. Interactions between proteins are presented as orange dots, and cells were counterstained with Phalloidin (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). For better illustration of the PLA signals, a ×20 magnification of the insets in Vac14 and Dock7 are also shown. (4) Quantitative analysis of the number of interactions per cell surface based on analysis of the PLA signals by Image Studio are presented in the bar graph. Comparisons were made against the negative control. Bars represent mean ± SEM. *P value < .05.
Dock7 and Vac14, but not Sec16a bind the BEACH domain
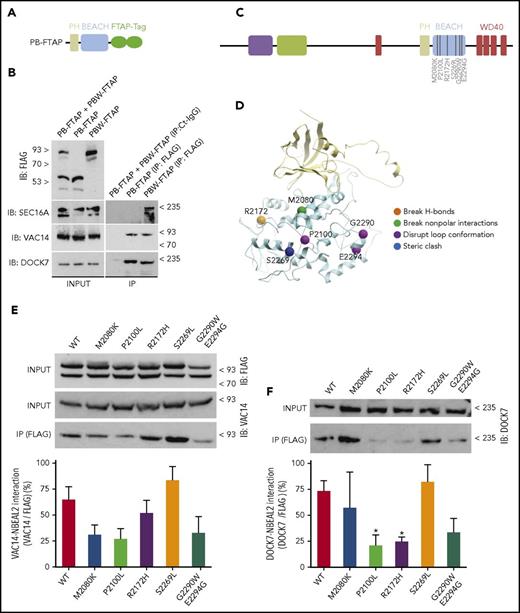
The BEACH domain intimately interacts with its adjacent aminoterminal PH domain over an interface of 1300 Å2, suggesting they may fold as a single functional unit.30,31 We therefore overexpressed a construct comprising both the PH and the BEACH domains (PB) in HEKs (Figure 3A). Dock7 and Vac14 were present in PB-FTAP and PBW-FTAP pull downs, whereas Sec16a was recovered only with PBW-FTAP (Figure 3B). Hence, the binding of Sec16a to Nbeal2 may be mediated through the WD40 domain, whereas the other 2 proteins bind to either the PH or the BEACH domain.
Effect of missense variants in the BEACH domain identified in GPS cases on the interaction with Dock7 and Vac14. (A) Diagram representing the construct PB-FTAP including the PH and BEACH domains of Nbeal2 and the FTAP-tag. (B) HEKs whole cell lysates containing a mixture of PB-FTAP and PBW-FTAP in a 1:1 ratio, or only PB-FTAP and PBW-FTAP (input) were immunoprecipitated using an isotype control IgG (Ct-IgG) or anti-FLAG. (C) Diagram representing the functional domains of Nbeal2 as per Figure 1A. Six amino acid mutations causative of GPS were selected on basis of their predicted functional consequences. (D) Computer model of the BEACH domain illustrating the 6 mutations shown in panel C and color-classified by disruptive effect. (E) PBW-FTAP in HEKs expressing the reference allele or the 6 mutations were lysed (input), IP with anti-FLAG and blotted with anti-Vac14. Quantitation of the interaction Vac14-Nbeal2 for each of the variants and control was achieved by densitometry. (F) Similar assays performed in panel E, but immunoblots (IBs) were developed with anti-Dock7. Experiments were performed on 5 independent occasions. Bars represent mean ± SEM. *P value < .05.
Effect of missense variants in the BEACH domain identified in GPS cases on the interaction with Dock7 and Vac14. (A) Diagram representing the construct PB-FTAP including the PH and BEACH domains of Nbeal2 and the FTAP-tag. (B) HEKs whole cell lysates containing a mixture of PB-FTAP and PBW-FTAP in a 1:1 ratio, or only PB-FTAP and PBW-FTAP (input) were immunoprecipitated using an isotype control IgG (Ct-IgG) or anti-FLAG. (C) Diagram representing the functional domains of Nbeal2 as per Figure 1A. Six amino acid mutations causative of GPS were selected on basis of their predicted functional consequences. (D) Computer model of the BEACH domain illustrating the 6 mutations shown in panel C and color-classified by disruptive effect. (E) PBW-FTAP in HEKs expressing the reference allele or the 6 mutations were lysed (input), IP with anti-FLAG and blotted with anti-Vac14. Quantitation of the interaction Vac14-Nbeal2 for each of the variants and control was achieved by densitometry. (F) Similar assays performed in panel E, but immunoblots (IBs) were developed with anti-Dock7. Experiments were performed on 5 independent occasions. Bars represent mean ± SEM. *P value < .05.
Impact of GPS-causing variants on the interaction of Dock7 and Vac14
To determine the possible effects of GPS-causing mutations, we cataloged all reported missense variants (n = 22)21,23,44,45 and those identified “in-house” by the ThromboGenomics41 and National Institute for Health Research (NIHR) BioResource sequencing projects (n = 14; Kate Downes, Kathleen Freson, and Keith Gomez, unpublished data [see acknowledgement]). This showed a significant fivefold enrichment (Fisher test, P value = 3.64 × 10−5) of variants in the BEACH domain, which harbored 13 of 36 of the observed variants (36%), whereas the domain only accounts for 292 of the 2754 (11%) amino acids of Nbeal2. We used the coordinates of the crystal structure of the BEACH domain of Lrba (pdb 1T77) to estimate the functional consequences of the 13 mutations and selected 6 with different types of structural changes to determine their effects on the binding of Dock7 and Vac14 (Figure 3C-D). HEKs were stably transfected with PBW-FTAP constructs and the reference, and 5 variant-carrying baits were expressed at similar levels, indicating a minimal, if any, detrimental effect of each variant on the folding of the domains (Figure 3E). Our results showed that M2080K, P2100L, and G2290W/E2294G (both on the same haplotype) seemed to diminish the binding of Vac14 (Figure 3E); P2100L and R2172H significantly reduced Dock7 binding, and its binding to the G2290W/E2294G bait seemed to be at a reduced level, although the difference with the reference bait was not significant (Figure 3F).
Localization of PBW-FTAP and Nbeal2 by electron and confocal microscopy
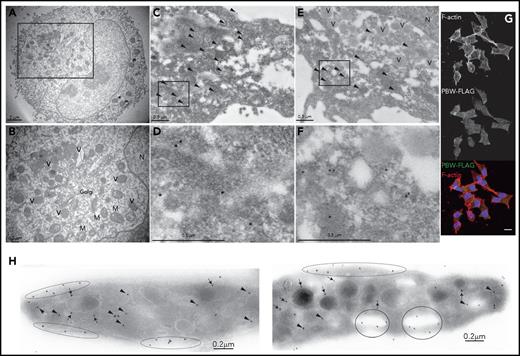
Currently, the subcellular localization of Nbeal2 remains unknown. We investigated this using PBW-FTAP CHRFs by labeling its FLAG epitope. Transmission electron microscopy revealed the presence of intracellular vesicles, mitochondria, and Golgi in CHRFs (Figure 4A-B). Labeling of FLAG by gold-adsorbed immunoglobulins did not show any staining in CHRFs (not shown). However, clusters of gold were observed across the cytoplasm of PBW-FTAP CHRFs, suggesting a cytosolic localization of PBW (Figure 4CF). This pattern was also confirmed by confocal microscopy by labeling PBW and F-actin with anti-FLAG and Phalloidin, respectively (Figure 4G; supplemental Figure 6). The use of a rabbit antiserum revealed the presence of Nbeal2 in the cytoplasm (arrowheads), in close proximity to enclosing membranes of the α-granules (arrows), the open canalicular system (OCS) (circles), and the cell surface (flat ellipses) of human platelets (Figure 4H).
Subcellular localization of PBW-FTAP and Nbeal2. (A) Ultrastructure of PBW-FTAP in CHRFs by transmission electron microscopy (TEM). (B) High magnification of the area squared in panel A showing intracellular vesicles (V), mitochondria (M), Golgi, and the nucleus (N). (C) Indirect gold-labeling of PBW-FTAP using anti-FLAG. Arrowheads point to clusters of gold across the cytoplasm of CHRFs. (D) High magnification of the boxed area in panel C. (E) PBW-FTAP in CHRFs stained as in panel C showing gold clusters in the cytoplasm but not in the intracellular vesicles. (F) High magnification of the boxed area in panel E. (G) Labeling of F-actin filaments with Phalloidin (top panel), PBW-FTAP (middle panel), and overlay (bottom panel) of the results from top and middle panels, visualized by confocal microscopy; nucleus stained with DAPI. Scale bar (which applies to all subpanels in panel G) represents 10 μm. (H) Gold-labeling of Nbeal2 in human platelets with rabbit antiserum. Gold particles label the platelet cytoplasm (arrowheads), α-granules (arrows), OCS (circles), and periphery (flat ellipses). Images are representative of the results of 2 TEM, 1 (gold-labeling), and 3 (confocal microscopy) independent experiments.
Subcellular localization of PBW-FTAP and Nbeal2. (A) Ultrastructure of PBW-FTAP in CHRFs by transmission electron microscopy (TEM). (B) High magnification of the area squared in panel A showing intracellular vesicles (V), mitochondria (M), Golgi, and the nucleus (N). (C) Indirect gold-labeling of PBW-FTAP using anti-FLAG. Arrowheads point to clusters of gold across the cytoplasm of CHRFs. (D) High magnification of the boxed area in panel C. (E) PBW-FTAP in CHRFs stained as in panel C showing gold clusters in the cytoplasm but not in the intracellular vesicles. (F) High magnification of the boxed area in panel E. (G) Labeling of F-actin filaments with Phalloidin (top panel), PBW-FTAP (middle panel), and overlay (bottom panel) of the results from top and middle panels, visualized by confocal microscopy; nucleus stained with DAPI. Scale bar (which applies to all subpanels in panel G) represents 10 μm. (H) Gold-labeling of Nbeal2 in human platelets with rabbit antiserum. Gold particles label the platelet cytoplasm (arrowheads), α-granules (arrows), OCS (circles), and periphery (flat ellipses). Images are representative of the results of 2 TEM, 1 (gold-labeling), and 3 (confocal microscopy) independent experiments.
Subcellular localization of Nbeal2, Dock7, Sec16a, and Vac14 in platelets
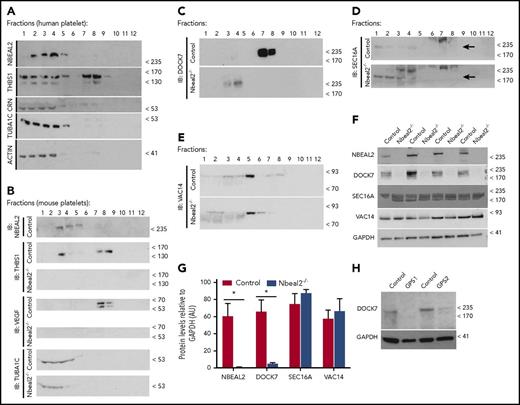
We further investigated the localization of Nbeal2 and its interactors Dock7, Sec16a, and Vac14 by subcellular fractionation of platelets. We first fractionated human platelets and identified cytoskeleton and dense tubular system in fractions 1 to 4 (enriched for tubulin, actin, and calreticulin) and α-granules in fraction 7 and 8 (enriched for the α-granule protein Thrombospondin-1 [Thbs1]) (Figure 5A). Nbeal2 was mostly found in fractions 2 to 4 containing cytoskeletal and dense tubular system proteins (Figure 5A). Fractionation of murine platelets showed a similar pattern (ie, Nbeal2 was primarily found in fractions 3-4) (Figure 5B). As expected, proteins typically stored in α-granules, Thbs1 and vascular endothelial growth factor, are significantly reduced in GPS platelets (Figure 5B). Dock7 was present in the α-granule-containing fraction (7-8) of control platelets but was mislocalized in fraction 4 in Nbeal2−/− platelets (Figure 5C). Although at lower levels, Dock7 and Nbeal2 colocalize in fractions 3 to 4 and 7 to 8, respectively, providing further evidence of their interaction (supplemental Figure 7). The localization of Sec16a and Vac14 did not differ significantly between platelets from both groups (Figure 5D-E), despite potential apparent increased levels of Sec16a in Nbeal2−/− platelets (Figure 5D), an observation that was not replicated in whole platelet lysates (Figure 5F-G).
Subcellular localization of Nbeal2 and its interactors, Dock7, Sec16a, and Vac14 in platelets. (A-B) SDS-PAGE separated fractions 1 to 12 obtained by sucrose gradient centrifugation of human (A) and murine (B) platelets probed in IB with specific antibodies against Nbeal2, Thbs1, Crn (Calreticulin), Tuba1c (Tubulin α 1c), and actin (β-actin); for the murine platelets, Crn was not probed but a specific antibody against Vegf (vascular endothelial growth factor) was included. (C-E) Localization of Nbeal2 interactors by IB in the different fractions obtained from platelets of control and Nbeal2−/− mice, (C) Dock7 in fraction 7 to 8 and fractions 3 to 4, respectively; (D) Sec16a in fraction 1 to 4; (E) Vac14 in fraction 5. (F) Levels of Nbeal2, Dock7, Sec16a, and Vac14 in platelets from control and Nbeal2−/− mice. (G) Densitometry of the western blot results shown in panel F; total lysates from panel F were performed on platelets of at least 4 (Nbeal2, Sec16a, Vac14) and 8 (Dock7) mice per genotype group. (H) Levels of Dock7 in platelets from GPS cases. For experiments in panels G and H, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control, and levels of the other proteins were normalized to GAPDH. Bars represent mean ± SEM. *P value < .05.
Subcellular localization of Nbeal2 and its interactors, Dock7, Sec16a, and Vac14 in platelets. (A-B) SDS-PAGE separated fractions 1 to 12 obtained by sucrose gradient centrifugation of human (A) and murine (B) platelets probed in IB with specific antibodies against Nbeal2, Thbs1, Crn (Calreticulin), Tuba1c (Tubulin α 1c), and actin (β-actin); for the murine platelets, Crn was not probed but a specific antibody against Vegf (vascular endothelial growth factor) was included. (C-E) Localization of Nbeal2 interactors by IB in the different fractions obtained from platelets of control and Nbeal2−/− mice, (C) Dock7 in fraction 7 to 8 and fractions 3 to 4, respectively; (D) Sec16a in fraction 1 to 4; (E) Vac14 in fraction 5. (F) Levels of Nbeal2, Dock7, Sec16a, and Vac14 in platelets from control and Nbeal2−/− mice. (G) Densitometry of the western blot results shown in panel F; total lysates from panel F were performed on platelets of at least 4 (Nbeal2, Sec16a, Vac14) and 8 (Dock7) mice per genotype group. (H) Levels of Dock7 in platelets from GPS cases. For experiments in panels G and H, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control, and levels of the other proteins were normalized to GAPDH. Bars represent mean ± SEM. *P value < .05.
Having identified the impact of the absence of Nbeal2 on the subcellular localization of Dock7, we then asked whether it had an effect on the overall cellular abundance of the 3 interactors. Similar levels of Sec16a and Vac14 were confirmed in platelets from both groups, but the Dock7 protein level was significantly reduced in Nbeal2−/− platelets (Figure 5F-G; supplemental Figure 8), and importantly, also in the platelets of 2 GPS cases (Pedigree C of the previous report21 ) (Figure 5H). Transcript levels of Dock7 in mouse MKs did not differ significantly between control and Nbeal2−/− mice (Guerrero et al27 and supplemental Table 3), and IBs with anti-Dock7 did not reveal signs of proteolysis or higher Dock7 levels in plasma (not shown), suggesting that the Nbeal2 protein is likely involved in its posttranslational modification and/or stability.
The Dock7 signaling pathway is altered in Nbeal2−/− platelets
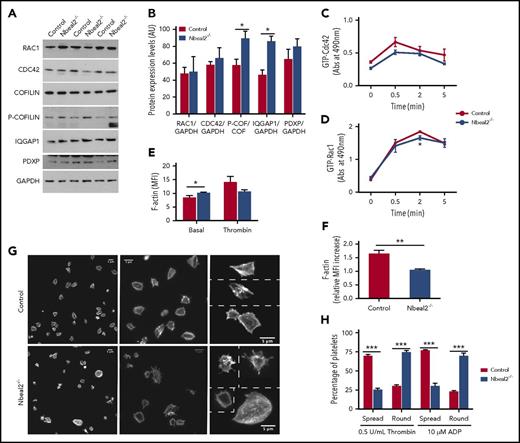
As a GEF, Dock7 exchanges GDP for GTP on Cdc42 and Rac1, causing their activation, followed by activation of downstream effectors, including the dephosphorylation (activation) of cofilin, a key regulator of actin turnover.46,47 Actin reorganization then leads to extension of filopodia and generation of lamellipodia,46 ultimately resulting in platelet shape change. Given the significant reduction of Dock7 levels and its altered localization in Nbeal2−/− platelets, we postulated that this canonical signaling pathway may be disrupted and set out to test this using control and Nbeal2−/− platelets.
First, the phosphorylated (inactive) form of cofilin, P-cofilin, was significantly increased in murine Nbeal2−/− platelets (Figure 6A-B). Other regulators of this signaling pathway such as Cdc42, Rac1, Iqgap1 and chronophin46,48 were also measured, and we found that the level of Iqgap1, a protein that stabilizes the active forms of Cdc42 and Rac1, was also significantly higher in Nbeal2−/− platelets (Figure 6A-B), suggesting that the increase in P-cofilin was likely due to an imbalance not only of Dock7 but also of Iqgap1. In fact, our previous gene expression array of control and Nbeal2−/− MKs revealed a significantly increased Iqgap1 transcript level in the latter (Guerrero et al 27 and supplemental Table 4), which is validated at the protein level in platelets in this study.
The Dock7 signaling pathway is altered in Nbeal2−/−platelets. (A) Platelet lysates from control and Nbeal2−/− mice were resolved by SDS-PAGE, and expression levels of Rac1, Cdc42, cofilin, P-cofilin, Iqgap1, Pdxp, and GAPDH were observed by IB with specific antibodies. (B) Densitometry of the results of the blots shown in panel A using Image Studio. (C-D) Time course of active Cdc42 (GTP-Cdc42) and active Rac1 (GTP-Rac1) in platelets stimulated with 0.5 U/mL thrombin. (E) F-actin levels in resting and thrombin-activated platelets. (F) Relative increase in F-actin levels in thrombin-activated vs resting platelets of control and Nbeal2−/− mice. (G) Representative images of phalloidin-labelled platelets spreading onto fibrinogen in response to thrombin from control and Nbeal2−/− mice at different magnifications. (H) Quantitation of fully spread and round platelets from control and Nbeal2−/− mice in response to activation with thrombin or adenosine 5′-diphosphate (ADP). Platelets in 5 random fields per condition were counted, with each field having between 80 and 120 platelets; 4 animals per genotype group were used. Bars represent mean ± SEM. MFI, mean fluorescence intensity. *P values < .05; **P values < .005; ***P values < .0005.
The Dock7 signaling pathway is altered in Nbeal2−/−platelets. (A) Platelet lysates from control and Nbeal2−/− mice were resolved by SDS-PAGE, and expression levels of Rac1, Cdc42, cofilin, P-cofilin, Iqgap1, Pdxp, and GAPDH were observed by IB with specific antibodies. (B) Densitometry of the results of the blots shown in panel A using Image Studio. (C-D) Time course of active Cdc42 (GTP-Cdc42) and active Rac1 (GTP-Rac1) in platelets stimulated with 0.5 U/mL thrombin. (E) F-actin levels in resting and thrombin-activated platelets. (F) Relative increase in F-actin levels in thrombin-activated vs resting platelets of control and Nbeal2−/− mice. (G) Representative images of phalloidin-labelled platelets spreading onto fibrinogen in response to thrombin from control and Nbeal2−/− mice at different magnifications. (H) Quantitation of fully spread and round platelets from control and Nbeal2−/− mice in response to activation with thrombin or adenosine 5′-diphosphate (ADP). Platelets in 5 random fields per condition were counted, with each field having between 80 and 120 platelets; 4 animals per genotype group were used. Bars represent mean ± SEM. MFI, mean fluorescence intensity. *P values < .05; **P values < .005; ***P values < .0005.
We then asked if the disequilibrium in the Dock7/Iqgap1/P-cofilin pathway had an effect on the function of platelets. First, levels of active Rac1 at 2 minutes after activation by thrombin were significantly higher (P value = .026) in platelets from control vs Nbeal2−/− mice, and a similar trend was observed for Cdc42, but this did not reach significance (Figure 6C-D). Second, we measured F-actin formation in platelets. Despite higher levels of F-actin in basal Nbeal2−/− platelets due to their larger size (Figure 6E; P value = .048), induction of actin polymerization by thrombin revealed a lower increase in Nbeal2−/− platelets (Figure 6E-F; P value = .0005). Last, we checked the endpoint of this pathway (ie, platelet shape change) by measuring the ability of platelets to spread onto fibrinogen after activation with both adenosine 5′-diphosphate and thrombin. Enumeration of activated vs nonactivated platelets assessed by shape change and spreading revealed that nearly all control platelets became activated, whereas the Nbeal2−/− platelets showed significantly reduced spreading (Figure 6G-H; P value ≤ .0005). Altogether, these results are compatible with the notion that Nbeal2 plays an important role in the regulation of cytoskeletal rearrangements in platelets, most likely through its interaction with Dock7.
Discussion
Despite the severity of inherited granule pathologies caused by mutations in NBEAL2, NBEA, and LYST, the key roles of the encoded BEACH proteins in the ontogeny of α-, δ-, and lysosomal granules remain poorly defined. Here, we report the results of studies aimed at defining the role of Nbeal2 at the biochemical and cellular level.
We identified the putative interactors of Nbeal2 in human cells and selected 10 candidate interactors for validation. The interactions of Dock7, Sec16a, and Vac14 with Nbeal2 were confirmed, and the physical proximity of the former 2 to Nbeal2 in human MKs was corroborated by PLA. Furthermore, the introduction of GPS-causing mutations in the BEACH domain showed profound effects on the interactions with Dock7. Although Nbeal2 is mostly present in the platelet cytoplasm, Dock7 localizes to the α-granules, and, interestingly, murine and human platelets devoid of functional Nbeal2 showed strongly reduced Dock7 levels, resulting in changes in its canonical signaling pathway.
The fivefold enrichment of GPS-causing variants in the BEACH domain prompted us to perform the pull-downs with a construct containing the BEACH and its PH and WD40 flanking domains. Despite the potential presence of false-positive interactors frequently seen in proteomics assays, 70% of the 129 proteins in Nbeal2’s interactome are present as primary nodes in the PPIN, which represents a lion’s share of the proteome of human MKs. So far, no similar interaction data are available for any of the BEACH proteins, except for the 21 proteins possibly interacting with Lyst.49
The observation that Nbeal2 localizes in the vicinity of the membrane, OCS, and α-granules of platelets is compatible with its assumed role in membrane dynamics, and especially α-granule trafficking. Our results are supportive of the notion that the GEF Dock7 may be a critical intermediary in this membrane dynamics, which is also essential in the formation of platelets by MKs.46 Therefore, the discovery that Dock7 is nearly depleted from platelets of GPS cases and Nbeal2−/− mice provides a plausible explanation as to why these platelets have an increased volume. In addition, our genome-wide association study identified variants associated with platelet traits in 9 genes encoding proteins in the Nbeal2 interactome (Nbeal2 itself, Dock7, and the 7 circular orange nodes with appended gene names in Figure 1E). The variant rs60286666 in DOCK7 is associated with the distribution width of platelet volume (P value = 8.59 × 10−33), providing further evidence of this Nbeal2 interactor possibly being important in the formation of platelets by MKs.39 The role of Dock7 in platelet function has not been studied, but the misty mouse, with a truncation in Dock7, has a prolonged bleeding time possibly explained by a reduced ability of platelets to support hemostasis.50 The relevance of the canonical Rac1/Cdc42/Dock7 signaling pathway in megakaryopoiesis and platelet function has however been extensively studied. The conditional deletion of cofilin51 and simultaneous deficiency of Rac1 and Cdc4252 result in decreased platelet count, larger platelets, and impairment of their function, including defective shape change. Moreover, dysregulation of cofilin was also observed in a mouse knock-in and a family with type 2B von Willebrand disease, both presenting with macrothrombocytopenia.53 Similar to these reports, we show that several elements of this signaling pathway (ie, P-cofilin and Iqgap1, a stabilizer of the active form of Cdc42 and Rac1) are dysregulated in murine Nbeal2−/− platelets, probably as a consequence of the reduced levels of Dock7. Hence, GTP-Rac1 levels are diminished in thrombin-stimulated Nbeal2−/− platelets, which show reduced actin polymerization and are unable to change shape efficiently (Figure 6D-H). Indeed, taking all the above evidence together, it is reasonable to assume that the macrothrombocytopenia in Nbeal2−/− mice and in GPS cases is in part explained by the specific loss of Dock7.
Dock7 interacts with the PH-BEACH domain, and this interaction seems to be diminished by all but 1 of the 6 GPS-causing mutations surveyed (Figure 3F). It has been suggested that Dock7 or 1 of its binding partners is localized in or on the surface of α-granules, and its PIP2/PIP3 binding domain makes the latter more likely.54 Our results are supportive of this assumption, and we reason that the interaction of Dock7 and α-granules is mediated through the BEACH domain.
Sec16a defines the budding sites on the endoplasmic reticulum membrane for trafficking of vesicles through the Golgi,55 and disruption of its binding to mutant WD40 domains (Figure 3B) could potentially affect platelet homeostasis. Similarly, changes in the function of Vac14, which in complex with Fig4 and Pikfyve (Figure 1H) regulates the levels of PI(3-5)P2, may alter endosomal trafficking and signaling.56
In summary, from 129 binding partners of Nbeal2 identified by mass spectrometry, we have confirmed the interaction of 3, Dock7, Sec16a, and Vac14, by different biochemical and cellular approaches and have also studied the functional relevance of 1, Dock7, in platelet biology and its involvement in the abnormal formation of platelets by MKs in GPS.
The mass spectrometry proteomics data reported in this article have been deposited in the PRIDE database of the ProteomeXchange Consortium (accession number PXD006091).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kate Downes (University of Cambridge, UK), Kathleen Freson (University of Leuven, Belgium), and Keith Gomez (University College London, UK) for reviewing the sequencing results.
Research in the Ouwehand laboratory is supported by grants from the European Commission (BLUEPRINT grant code HEALTH-F5-2011-282510, HORIZON 2020 grant code 692041), NIHR (for funding of the Cambridge Biomedical Research Centre, grant RG64219), Medical Research Council grant WT098503, Bristol Myers-Squibb, BHF grants RP-PG-0310-1002 and RE/13/6/30180 for the BHF Cambridge Centre of Excellence, and National Health Service Blood and Transplant (NHSBT). The sequencing of the cases with GPS was supported by the NIHR BioResource grant RBAG163. T.K.B. is the recipient of a Clinical Research Training Fellowship award from the British Society for Haematology and NHSBT, J. Collins is a Medical Research Council Clinical Training Research fellow, and W.H.O. is an NIHR Senior Investigator.
Authorship
Contribution: L.M., M.J., S.A.d.H., and J.A.G. designed and performed experiments and analyzed data; M.P., L.Y., and J. Choudhary carried out proteomics analysis; J. Collins, T.K.B., L.G., R.P., S.M., P.A.S., and W.J.A. analyzed data; P.N. performed EM of platelets; R.F. provided GPS samples; W.W.Y. carried out structural analysis; and W.H.O. and J.A.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.P., L.Y., and J.C. is The Institute of Cancer Research, Cancer Biology Division, Chester Beatty Laboratories, London, United Kingdom.
The current affiliation for S.M. is European Molecular Biology Laboratory European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, United Kingdom.
Correspondence: Jose A. Guerrero, Department of Haematology, University of Cambridge, Long Road, CB2 0PT, Cambridge, UK; e-mail: jg652@medschl.cam.ac.uk.

![Figure 1. The Nbeal2 interactome. (A) Diagram showing the functional domains of Nbeal2 (ARM, Armadillo; ConA, Concanavalin A; WD40 repeat [WD40]). Construct 1 (PBW-FTAP) encompasses the PH, BEACH, and aminoterminal WD40 domains and the FTAP tag. Construct 2 (GFP-FTAP) encompasses GFP and FTAP. (B) Ectopic expression of GFP-FTAP and PBW-FTAP in HEKs. (C) FTAP-purification of GFP-FTAP and PBW-FTAP from HEKs. Purified proteins were resolved by SDS-PAGE, visualized by Coomassie blue stain, and analyzed by liquid chromatography-mass spectrometry. Some lanes in gels from panels B and C were deleted and replaced by black vertical lines. (D) Identified proteins (n = 129) were classified according to their protein class using gene ontology terms. (E) PPIN; 637 bait proteins representing the processes of hemostasis, megakaryopoiesis, and platelet formation were used to retrieve 5112 nodes (proteins) and 26 702 edges (biochemical reactions) by Reactome and IntAct. Orange nodes (n = 91) are interactors present in the PPIN, and the purple nodes (n = 38) have interactions with 1 or more proteins of the PPIN. Nbeal2 is shown in black and Dock7, Sec16a, and Vac14 are shown as orange triangles. (F-H) Subnetworks extracted from the PPIN in panel E for Dock7 (F), Sec16a (G), and Vac14 (H). The expression levels for the corresponding genes determined by the sequencing of RNA of human MKs and expressed as log2 FPKM (fragments per kilobase of transcript per million mapped reads) is presented in white-to-green color, and only nodes corresponding to genes with FPKM > 1 in MKs are depicted (see color bar for relative FPKM values).43 The gray/black (E-H), light blue (F-H), and dark blue (F-H) edges are based on the results of pull-down experiments reported in this study, Reactome and IntAct, respectively. A Cytoscape file to generate an interactive version of the PPIN, containing gene expression levels and other annotation features, is available from the supplemental data. MW, molecular weight.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/9/10.1182_blood-2017-08-800359/4/m_blood800359f1.jpeg?Expires=1770950169&Signature=y8LaGk6D0zHkSYL1xdkjJubpMVk0Iaqpx6K93hfTZ~v89adaZeoCEUMbpGsky9xYelX6-gUuiYVdRGTa5Fgzc1dsfVrVs4EL0fIJANGJSMA-9rWDG5tvHxZwKddL7MFYD3GR6~mbpPLgJ8ZW--roypbkTrF4JO9AV8ICkPtuKcIR1V-hkIE3JaH~ZDltGmqSxeCMmyCfrelFfflcaHoLn2CzwpIZ~1u0C-TYlgxAY5fr0QjMpwlxckE7KYS8iY-JOvFER8gRWVuueGYk0ej7PWctA9BvkdbtMb6JJ6sYr791ZrJwuwHxvBxMaH0CJkKV~Vo44pmZYwrlCVhminYOig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal