In this issue of Blood, Kohnken et al demonstrate a novel oncogenic pathway featuring IL-15-miR-29b-BRD4 positive feedback loop and targeting bromodomain-containing protein 4 (BRD4) disables the oncogenic loop, identifying a potentially effective target for therapy for cutaneous T-cell lymphoma (CTCL) patients.1
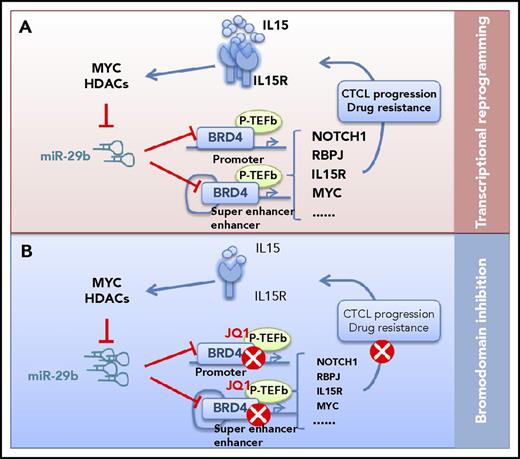
A novel oncogenic pathway and potential targets for CTCL. (A) IL-15 represses miR-29b levels through recruitment of Myc/Hdac-1 at the miR-29b regulatory region in CTCL cells. The decreased miR-29b releases its negative regulation on BRD4, which then binds acetylated lysine residues on chromatin at promoter and superenhancer of oncogenic genes. BRD4 regulates tumor driver genes such as NOTCH1, MYC, and RBPJ, coordinates transcriptional reprogramming, and promotes CTCL progression and drug resistance. (B) Bromodomain inhibition (via JQ1) can block dynamic transcriptional responses, remodeling the enhancers and key signaling outputs required for CTCL to progress.
A novel oncogenic pathway and potential targets for CTCL. (A) IL-15 represses miR-29b levels through recruitment of Myc/Hdac-1 at the miR-29b regulatory region in CTCL cells. The decreased miR-29b releases its negative regulation on BRD4, which then binds acetylated lysine residues on chromatin at promoter and superenhancer of oncogenic genes. BRD4 regulates tumor driver genes such as NOTCH1, MYC, and RBPJ, coordinates transcriptional reprogramming, and promotes CTCL progression and drug resistance. (B) Bromodomain inhibition (via JQ1) can block dynamic transcriptional responses, remodeling the enhancers and key signaling outputs required for CTCL to progress.
CTCL is a non-Hodgkin lymphoma in which malignant CD4+ T cells home to skin causing a variety of clinical manifestations ranging from mycosis fungoides (characterized by localized skin tumors) to leukemic CTCL (Sézary syndrome), where malignant T cells invade the peripheral lymphocyte compartment.2 In the early stages, patients often have skin-restricted disease, and in advanced stages, CTCL is a fatal disease3 with involvement of the lymph node, viscera, and/or blood compartments. CTCLs are believed to originate from skin-tropic mature CD4+ T cells.4 Despite the development of new therapeutic agents, there is no curative treatment. Drug resistance is a common problem, and patients with advanced-stage disease have very poor clinical outcomes. Thus, there is an urgent unmet need to understand the molecular pathways that sustain the neoplastic phenotype in CTCL and identify CTCL-specific pathways that would predict vulnerability to therapies aimed at those pathways.
Recent studies,5 notably via exome sequencing and expression analysis, have made significant progress in understanding the molecular determinants of CTCL. These analyses have identified a number of key genes and signaling pathways, whose frequent amplification and constitutive activation, or loss, suggested a pathogenic role in advanced CTCL. Frequently amplified genes include MYC, STAT3, and STAT5B, whereas frequently lost genes include TP53, PTEN, and CDKN1A/B. In addition, recurrent genomic gains at loci for genes encoding cytokines and their receptors (IL-2, IL-7) and loss of negative regulators of cytokine transcription (TCF8/ZEB1, DUSP5) highlighted the importance of aberrant cytokine signaling in the pathogenesis of CTCL.
Among the dysregulated cytokines, interleukin-15 (IL-15) appears to be essential for lymphoma cell growth and plays a key pathogenic role in CTCL.6 Strong IL-15 expression is a characteristic feature of CTCL.7 In CTCL, IL-15 has been implicated in the recruitment of CD4+ memory T cells to the skin, induction of T-cell proliferation, and inhibition of apoptotic cell death.8 Recently, Mishra et al confirmed the role of IL-15 by demonstrating stage-dependent IL-15 overexpression of CD4+ T cells from patients with CTCL, and the CTCL in IL-15 transgenic mice closely resembling human CTCL with shared molecular aberrations. They provided in vivo evidence that IL-15 has a causal role in the pathogenesis of CTCL, in part via the epigenetic inhibition of the transcriptional repressor ZEB1, which in turn leads to overexpression of IL-15 and activation of specific histone deacetylase (HDAC).9
To gain insight into mechanisms of IL-15 overexpression and its role in CTCL pathogenesis, Kohnken et al characterized the role of microRNAs (miRs) in epigenetic modifier-dependent transcriptional regulation in IL-15 expression. Using patient CD4+ T cells, the group revealed low levels of miR-29b, an inverse relationship between miR-29b and BRD4, and overexpression of BRD4 in CTCL cells. Chromatin immunoprecipitation and sequencing analysis (ChIP) showed increased genome-wide BRD4 occupancy at promoter and enhancer regions in CD4+ T cells from CTCL patients, leading to increased expression of oncogenic driver genes such as NOTCH1 and RBPJ. ChIP-seq analysis also found that BRD4 binding is increased in CTCL patient cells at promoter regions of all 3 components of the IL-15 receptor complex, IL-15Rα, IL-15Rβ, and IL-15Rγ, which enhance IL-15 autocrine signaling. Next, using an in vivo IL-15 transgenic mouse model of CTCL, the authors demonstrated that inhibiting BRD4 binding via JQ1 treatment prevented progression of CTCL, thus supporting a novel oncogenic pathway that includes IL-15, miR-29b, and BRD4 in CTCL and suggesting therapy for CTCL patients. To examine the role of this pathway in CTCL, IL-15 treatment significantly decreases the pri-miR-29b1 level in CTCL cells compared with untreated cells, whereas BRD4 protein expression was increased. Consistently, IL-15 transgenic mice have increased expression of BRD4, NOTCH, and RBPJ. Together, these data show that IL-15 repressed miR-29b levels in CTCL cells through recruitment of Myc/Hdac-1 at the miR-29b regulatory region and that repressed miR-29b level contributes to increased BRD4 activity, IL-15 signaling, and ultimately, CTCL disease pathogenesis (see figure).
The role of epigenetic dysregulation in the pathogenesis of CTCL is an area of active research and clinical interest. In addition to global changes such as promoter DNA hypomethylation, mutations in epigenetic modifiers have been identified in CTCL patients and suggest they play a role in the development of disease. Epigenetic reader protein BRD4 has been a therapeutic target of interest in several tumor types. In this report by Kohnken et al, the authors demonstrated IL-15 signaling and downstream miR-29b as novel regulators of BRD4 protein expression in CTCL. The authors described the efficacy of JQ1 as an antitumor agent in CTCL, which acts by inducing cell-cycle arrest in cell lines and preventing disease progression in IL-15 transgenic mice. The significance of these findings is highlighted by demonstration of sustained oncogenic signaling through positive feedback IL-15-miR29b-BRD4 signaling loop and the extensive binding activity of BRD4 at regulatory regions in CTCL cells. Intriguingly, this binding profile can be reversed to that of normal T cells with JQ1 treatment. Indeed, molecular targeting of BRD4 by JQ1 and other bromodomain and extra-terminal (BET) inhibitors has been investigated as a treatment strategy in many hematologic malignancies.
Moreover, previous studies have identified that BRD4 binds chromatin in enhancer and superenhancer regions to direct downstream gene expression through interaction with transcription cofactors such as Mediator and p-TEFb in a variety of cancers.10 Because a large portion of BRD4 binding was detected in promoter and superenhancer regions and high expression of BRD4 is present in advanced CTCL, we speculated that BRD4 functions as a central hub to regulate tumor driver genes such as NOTCH1, MYC, and RBPJ, coordinating transcriptional reprogramming and promoting CTCL progression and drug resistance (see figure). Thus, BET inhibition can block dynamic transcriptional responses, remodeling the enhancers and key signaling outputs required for CTCL to progress.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal