TO THE EDITOR:
Addition of the CD33-targeted immunoconjugate gemtuzumab ozogamicin (GO; Pfizer) has been shown to improve the response to standard-induction chemotherapy and results in better long-term survival in adult patients with acute myeloid leukemia (AML).1 The greatest impact was observed in those with favorable-risk cytogenetics, with a lesser but still significant benefit in patients with intermediate-risk cytogenetics but no benefit in those with adverse-risk cytogenetics.1 Several studies have also demonstrated that the response positively correlates with higher levels of membrane CD33 expression on leukemic blasts.2-5 Data recently published by Lamba and colleagues further suggests that genotype at a common single nucleotide polymorphism (SNP) in the CD33 gene (rs12459419 C>T) determines response to GO in patients aged 0 to 29 years with de novo AML treated on the randomized phase 3 Children’s Oncology Group trial AAMLL0531.6 The SNP influences alternative splicing at CD33 exon 2 such that the C allele leads to expression of the full-length protein but the T allele is associated with increased levels of a truncated isoform lacking the external GO-binding domain. The authors found that only those patients with a homozygous CC genotype (∼50% of patients) had a favorable response to GO, with no clinical benefit in those with either the heterozygous CT or homozygous TT genotype. The impact of GO was greatest in the CC patients with favorable risk defined as favorable cytogenetics or the presence of NPM1 or CEBPA mutations. These data have important implications for the use of GO in AML, and are particularly pertinent in view of the recent approval by the US Food and Drug Administration of Mylotarg for treatment of AML. We therefore investigated whether similar results pertained to younger adult patients treated on UK Medical Research Council (MRC) AML15 (ISRCTN17161961) and National Cancer Research Institute (NCRI) AML17 (ISRCTN55675535) trials. Treatment protocols and outcomes were as reported previously.7,8 Informed patient consent was obtained in accordance with the Declaration of Helsinki, and ethical approval for tissue use was obtained from the Wales Research Ethics Committee 3.
Genomic DNA was available from 536 of 2063 patients who were entered into different GO randomizations in these trials, and a flowchart of patients studied is shown in supplemental Figure 1 (available on the Blood Web site). Of these, 25 patients were randomized to receive GO in induction and consolidation and 260 in induction alone; 218 were randomized to no-GO and 33 to receive GO in consolidation alone. The latter were included in the no-GO group for the analysis as there was no evidence of a benefit for GO in consolidation.7 There was no difference in overall survival (OS) between those who were included or not included in our study (P = .06), nor between those who were in different trials (P = .6). DNA was also available from a further 184 patients scheduled (not randomized) to receive GO.
Samples were screened for the CD33 SNP using HaeIII restriction enzyme digestion of polymerase chain reaction–generated amplicons (see supplemental Data). Genotype distribution was comparable to that observed by Lamba et al6 : 336 were CC (47%), 319 CT (44%), and 65 TT (9%), and the minor allele frequency was 30%. There were no differences in baseline characteristics between the genotypic groups, including age, sex, diagnosis (primary/secondary disease), World Health Organization (WHO) performance status, presenting white cell count, and cytogenetics (supplemental Table 1). The proportion of patients who received GO did not differ significantly according to genotype (52% of CC, 53% of CT, 60% of TT). The expression level of CD33 as evaluated by quantitative flow cytometry of CD33+ blasts had previously been reported on 249 of the above patients,5 and the median CD33 mean fluorescence intensity was 10.7 for CC genotype patients (range, 0.2-298.1), 11.1 for CT patients (range, 0.1-134.8), and 3.8 for TT patients (range, 0.1-13.3) (P = .0001 across all 3 groups) (supplemental Figure 2). This finding of a similar level of expression in the CC and CT groups but a much lower level in the TT group is in accord with the data of Lamba et al.6
In the randomized cohort of 536 patients, the 5-year relapse-free survival (RFS) and OS were similar in both arms (39% vs 42% and 46% vs 47% for GO vs no-GO, respectively, both P = .9). There was, however, a strong trend to a better outcome with GO in those patients with favorable cytogenetics (hazard ratio, 0.59; 95% confidence interval [CI], 0.30-1.14; P = .1 for RFS, and hazard ratio, 0.47; 95% CI, 0.22-1.01; P = .05 for OS). This preferential impact of GO in patients with favorable cytogenetics is in agreement with previous publications.1
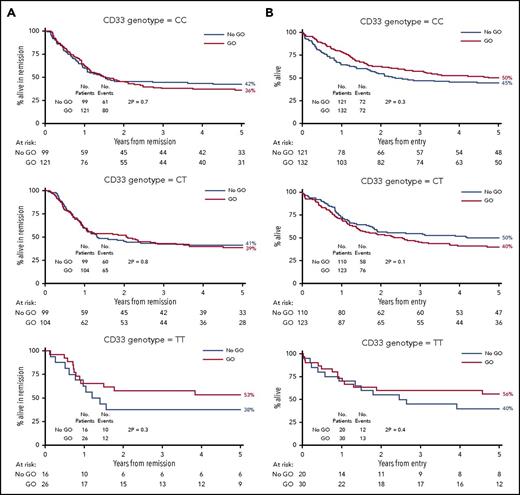
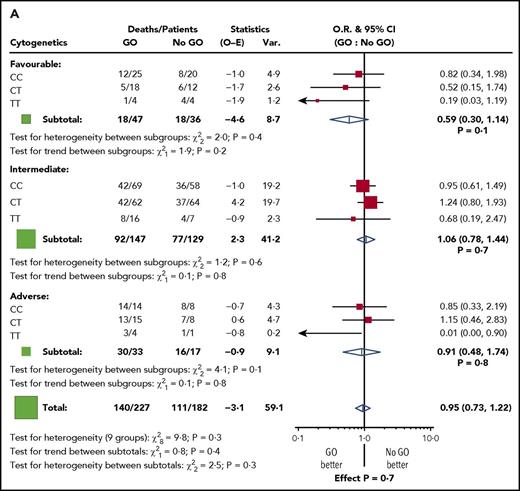
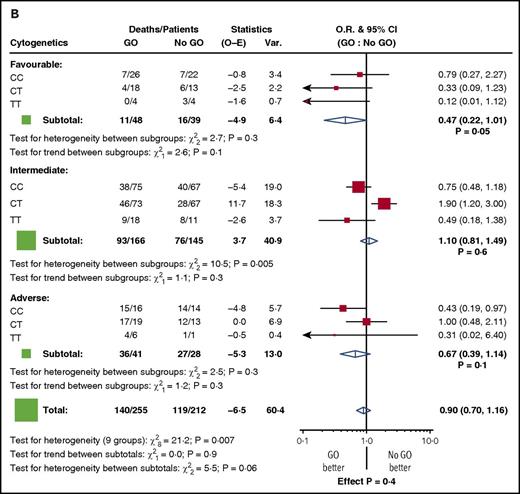
Among the randomized patients, we found no difference in response to GO in the genotype groups. Five-year RFS for GO vs no-GO was 36% vs 42% for CC patients (P = .7), 39% vs 41% for CT (P = .8), and 53% vs 38% for TT (P = .3) (Figure 1A). Similarly, 5-year OS was 50% vs 45% for CC patients (P = .3), 40% vs 50% for CT (P = .1), and 56% vs 40% for TT (P = .4) (Figure 1B). When the analysis was restricted to the 87 patients with favorable cytogenetics, there was again no discernible impact of the genotype (test for heterogeneity between subgroups: χ2, 2.0; P =.4 for RFS, and χ2, 2.7; P = .3 for OS) (Figure 2). In addition, there was no difference in the results according to the dose of GO administered (3 mg/m2 or 6 mg/m2, 152 vs 148 patients, respectively) (supplemental Figure 3).
Outcome according to CD33 genotype for SNP rs12459419 in 536 patients randomized to receive or not receive GO. (A) RFS. (B) OS.
Outcome according to CD33 genotype for SNP rs12459419 in 536 patients randomized to receive or not receive GO. (A) RFS. (B) OS.
Stratified analyses for outcome by cytogenetic risk group for patients in the GO randomization. (A) RFS. (B) OS. O-E, observed-expected; O.R., odds ratio; Var., variance.
Stratified analyses for outcome by cytogenetic risk group for patients in the GO randomization. (A) RFS. (B) OS. O-E, observed-expected; O.R., odds ratio; Var., variance.
It is difficult to explain why our results should differ so greatly from those of Lamba and colleagues. The genotype frequencies in the 2 populations were similar, as was the correlation between genotype and CD33 expression. Our patients were adults (age range, 13-69 years) whereas the patients in Children’s Oncology Group trial AAMLL0531 were mainly children (0-29 years). It is not obvious why a difference in patient age should have such an impact, although 1 possibility is that multidrug resistance due to P-glycoprotein–mediated drug efflux, which is higher in older patients9 and has been reported to influence response to GO,2 may mitigate against any benefit from the CC genotype in adult patients; this requires further investigation. The design of the randomized trials is also different, with varying schedules and doses used in the AML15 and AML17 trials investigated here, but a meta-analysis of adult patients did not suggest that these differences significantly impact outcome.1 Our study is limited by its size (536 patients randomized), but even if the number of patients were doubled, the chance of the GO effect being significantly greater only in the CC genotype group is <1 in 1000. Our findings are disappointing as the ability to predict a response to GO would have a major impact on patient management and would be cost-saving. Further studies of other randomized trials of GO addition to standard therapy, both in children and in adults, are warranted.
The online version of this article contains a data supplement.
Acknowledgments
These trials were registered at the UK Medical Research Council (MRC) as #ISRCTN17161961 and at the UK National Cancer Research Institute (NCRI) as #ISRCTN55675535.
This work was supported by Leukaemia and Lymphoma Research, now called Bloodwise, and was undertaken at University College London (UCL), which receives a proportion of funding from the UK Department of Health’s National Institute for Health Research (NIHR) Biomedical Research Centre’s funding scheme.
Authorship
Contribution: R.E.G. and T.P. performed genotyping; N.K., S.D.F., A.K.B., and N.H.R. provided data; M.W. and R.K.H. performed statistical analysis; R.E.G. and D.C.L. designed the study and wrote the paper; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, UCL Cancer Institute, 72 Huntley St, London WC1E 6DD, United Kingdom; e-mail: rosemary.gale@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal