Key Points
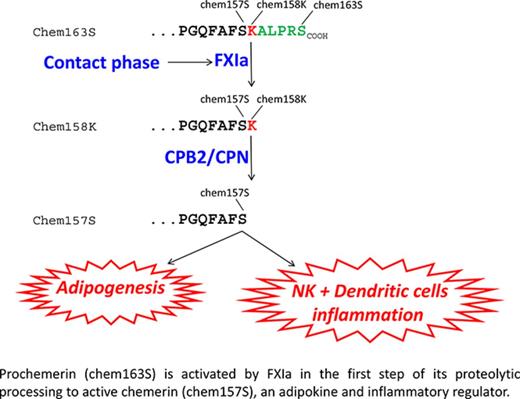
Inactive prochemerin is activated in plasma by coagulation enzymes to active chemerin forms that are adipokines and chemoattractants.
FXIa cleaves prochemerin, forming a partially active intermediate that is then fully activated by plasma basic carboxypeptidases.
Abstract
Chemerin is a chemoattractant and adipokine that circulates in blood as inactive prochemerin (chem163S). Chem163S is activated by a series of C-terminal proteolytic cleavages resulting in diverse chemerin forms with different levels of activity. We screened a panel of proteases in the coagulation, fibrinolytic, and inflammatory cascades to identify those that process prochemerin in plasma. Factor XIa (FXIa) cleaved chem163S, generating a novel chemerin form, chem162R, as an intermediate product, and chem158K, as the final product. Processing at Arg162 was not required for cleavage at Lys158 or regulation of chemerin bioactivity. Contact phase activation of human platelet-poor plasma by kaolin led to cleavage of chem163S, which was undetectable in FXI-depleted plasma and markedly enhanced in platelet-rich plasma (PRP). Contact phase activation by polyphosphate in PRP resulted in 75% cleavage of chem163S. This cleavage was partially inhibited by hirudin, which blocks thrombin activation of FXI. After activation of plasma, levels of the most potent form of chemerin, chem157S, as well as inactive chem155A, increased. Plasma levels of chem163S in FXI-deficient patients were significantly higher compared with a matched control group (91 ± 10 ng/mL vs 58 ± 3 ng/mL, n = 8; P < .01) and inversely correlated with the plasma FXI levels. Thus FXIa, generated on contact phase activation, cleaves chem163S to generate chem158K, which can be further processed to the most active chemerin form, providing a molecular link between coagulation and inflammation.
Introduction
The contact phase of the blood coagulation cascade consists of high–molecular-weight kininogen (HK) and the protease zymogens, factor XII (FXII), FXI, and prekallikrein (PK).1 Exposure of blood to negatively charged surfaces leads to activation of FXII to the protease FXIIa, which in turn catalyzes the activation of FXI to FXIa, leading to sequential formation of FIXa, Fxa, and subsequent thrombin generation constituting the intrinsic pathway.
Interest in the contact phase system has been renewed by the demonstration that polyphosphate (Polyp), a linear, highly anionic polymer released from either microbial pathogens or activated platelets, is a potent activator of the contact pathway.2 In addition, Polyp accelerates FXI activation by thombin, supporting the thesis that feedback activation of FXI by thombin represents an amplification pathway to augment thrombin generation.3,4
The contact phase also participates in thrombosis and inflammation.5,6 FXII deficiency in mouse models confers protection from pathological arterial thrombosis, but does not affect hemostasis.7,8 Inhibition of FXIa improves survival in a mouse sepsis model through blocking the coagulation cascade and altering cytokine levels, such as tumor necrosis factor-α, interleukin-6 (IL-6), and IL-10.9-12 FXI antisense oligonucleotide has demonstrated clinical efficacy in preventing postoperative venous thrombosis.13
Chemerin (retinoic acid receptor responder gene 2) is a chemoattractant and adipokine that was first discovered in psoriasis.14 Chemerin is secreted into blood as a 143–amino acid–inactive precursor, prochemerin (chem163S), undergoes precise proteolysis, mediated by a variety of enzymes involved in coagulation, fibrinolytic, and inflammatory cascades, in its C-terminus, leading to its sequential activation and inactivation. Plasmin cleaves chem163S at Lys158 to generate chem158K, which has low bioactivity, with subsequent cleavage by the basic plasma carboxypeptidases, carboxypeptidase N (CPN) or carboxypeptidase B2 (CPB2), to produce chem157S, the most active form of chemerin.15 Inactivation of chem157S to chem155A and smaller forms of chemerin occurs by further proteolytic cleavages.
Chemerin has 3 known receptors, 2 signaling G protein–coupled receptors, chemokine-like receptor 1 (CMKLR1), and G-protein receptor-1 plus a nonsignaling receptor C-C chemokine-like receptor 2f.14,16-18 CMKLR1 is extensively expressed on adipocytes and immune cells, including dendritic cells, natural killer cells, and macrophages,19-22 whereas G-protein receptor-1 is present on adipocytes, skeletal muscle cells, and brain cells.22,23
As an adipokine, chemerin regulates adipogenesis and adipocyte metabolism,24 promoting adipogenesis via CMKLR1 signaling, accompanied by an increase in the adipogenic markers, adiponectin and peroxisome proliferator-activated receptor γ. Peroxisome proliferator-activated receptor γ in turn drives chemerin expression in adipose tissue to form a positive feedback loop.25,26 Circulating levels of chemerin are elevated in obesity and metabolic syndrome.27 We recently reported that chemerin is activated in adipose tissue of obese subjects, with extensive C-terminal processing, resulting in substantial levels of novel degraded forms in plasma that correlate with obesity.28
Most of the chemerin in normal human plasma is inactive chem163S.29 To identify the physiological activator of chemerin in plasma, we screened proteases from the coagulation, fibrinolytic, and complement pathways. We found that FXIa was essential to generate active chemerin from chem163S in plasma, thus suggesting a new molecular link between thrombosis, inflammation, and adipogenesis.
Materials and methods
Extended Materials and methods are in the supplemental Methods, available on the Blood Web site.
Plasma samples
Patients and age- and sex-matched volunteers (supplemental Table 1) gave informed consent under protocols approved by Stanford University Medical Center and Partners Healthcare Institutional Review Boards or the Ethical Committee of the University of Perugia. FXI levels in plasma from FXI-deficient patients and controls were measured on an ACL Futura Plus coagulometer (Instrumentation Laboratories, Milan, Italy) using HemosIL reagents (Instrumentation Laboratory, Milan, Italy).
Production of human chemerin forms
Chemerin forms were produced and purified as described previously (supplemental Table 2).15 The purified proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The molecular mass and purity of the protein was confirmed by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry with internal standards of 2466, 3660, and 5734 Da included in the analysis (PAN Facility, Stanford University).
In vitro enzyme cleavage assays
Chem163S (10 μM) was incubated with 100 nM enzymes at 37°C for 30 minutes in phosphate-buffered saline containing 2 mM calcium chloride and 0.1 mM zinc chloride, and the reaction was terminated by dithiothreitol (10 mM) before analysis by SDS-PAGE and mass spectrometry.
FXIa cleavage of 6his-chem163S
FXIa (100 nM) and 6his-chem163S (10 μM) were incubated in FXIa buffer (50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 125 mM sodium chloride, 5 mM calcium chloride, and 0.1 mg/mL bovine serum albumin; pH 7.4) at 37°C for 30 minutes. Reactions were terminated by the addition of 100 nM of D-phenylalanyl-prolyl-arginyl chloromethyl ketone (PPACK), and products were analyzed by MALDI-TOF mass spectrometry. The amounts of 6his-chem163S consumption and 6his-chem158K generation were determined by chem163S and chem158K enzyme-linked immunosorbent assays (ELISAs) and western blots developed with horseradish peroxidase–conjugated goat anti-rabbit antibody (100 ng/mL) and detected with enhanced chemiluminescence film (GE Healthcare).
Specific ELISAs for chemerin forms
Kinetic analysis of FXIa cleavage of chemerin peptides and full-length protein
To determine the kinetic constants for cleavage of chemerin and FIX by FXIa, peptides (1 μM to 3 mM, supplemental Table 3) representing the C-terminus and the FXIa cleavage site in FIX were treated with FXIa (30 nM) in FXIa assay buffer for 30 minutes at 37°C before terminating the reaction with 100 nM PPACK. A total of 100 μl of each reaction mixture was applied to a Zorbax Eclipse Plus C18 (4.6 × 150 mm) column (Agilent, Santa Clara, CA) and separated with a 0% to 40% acetonitrile gradient in 0.1% trifluoroacetic acid (volume-to-volume ratio) by high-performance liquid chromatography (HPLC). The concentration of peptides present in the reaction mixture was determined from a standard curve of that peptide. The standard curves were constructed with peptides (1 μM to 3 mM).
For the protein, chem163S (0.33-1000 nM) was incubated with FXIa (30 nM) in assay buffer at 37°C for 30 minutes. The reaction was stopped by 100 nM PPACK. Generation of chem158K was measured by specific ELISA.
The values for Km and kcat were determined by fitting to the Michaelis-Menten equation by nonlinear regression using Prism version 6 (GraphPad, San Diego, CA). Experiments were performed 3 times independently, and the data were pooled for analysis.
Preparation of platelet-rich plasma and platelet-poor plasma
Blood was drawn into 3.8% sodium citrate (BD Biosciences) and platelet-rich plasma (PRP) prepared by centrifugation at 250 × g for 10 minutes at room temperature. Platelet-poor plasma (PPP) was prepared by centrifugation of the PRP at 1200 × g for 10 minutes at room temperature.
Cleavage of chemerin in PPP, PRP, and FXI-depleted plasma
For PPP, MP reagent (5 μM; Diagnostica Stago, Parsippany, NJ), GPRP (5 mM), kaolin (5 μg/mL), 6his-chem163S (10 μM), and calcium (Ca++) (5 mM) were incubated in human pooled normal plasma (George King Bio-medical) or FXI-depleted plasma (Heamatologic Technologies) at 37°C. For PRP, GPRP (5 mM), kaolin (5 μg/mL), or polyP (5 μM) and Ca++ (5 mM) were incubated at 37°C for 30 minutes. In some experiments, hirudin (1.5 U/mL) was added to block thrombin activity30 in PolyP-triggered PRP. All reactions were terminated by the addition of PPACK (100 μM) at various time points up to 30 minutes.
Preparation of plasma for chemerin form ELISAs
Plasma, before and after contact phase activation with either kaolin or Ca++-saturated polyP (30 to >500 phosphate units; Sigma, St. Louis, MO), was mixed with 100 μl of heparin-agarose (Sigma) and complete protease inhibitor (Roche Applied Science). After incubation at 4°C for 2 hours, the heparin-agarose beads were pelleted and washed extensively with PBS, and chemerin was eluted with 0.8 M sodium chloride in PBS, all in the presence of complete protease inhibitor. The eluted proteins were diluted with 1% bovine serum albumin in phosphate-buffered saline for analysis by the specific chemerin form ELISAs.
FXIa fluorogenic assay
MP reagent (5 μM), GPRP (5 mM), kaolin (5 μg/mL), and 50 μM FXIa fluorogenic substrate D-LPR-ANSNH-C3H7•2HCl (Haematologic Technologies) were diluted in plasma at 37°C. The reactions were initiated by recalcifying either PRP or PPP with 5 mM Ca++, and the fluorescence intensity was monitored at excitation/emission wavelengths of 352/470 nm using a FLUOROSKAN ASCENT FL fluorescent plate reader (Thermo Electron). The assay was repeated ≥4 times independently. Calibration curves were constructed with FXIa (range, 0.1-100 nM).
Calcium mobilization assay
Calcium mobilization was determined in cells expressing human CMKLR1 (hCMKLR1/L1.2 cells)15
Statistics
Comparison of 2 groups was by Student t test; multigroup comparisons were by 2-way analysis of variance followed by Tukey correction for multiple testing. The correlation analysis was calculated with 2-tailed Pearson correlation coefficients. Analyses were performed using Prism version 6. P < .05 was considered significant.
Results
Protease screening of prochemerin cleavage
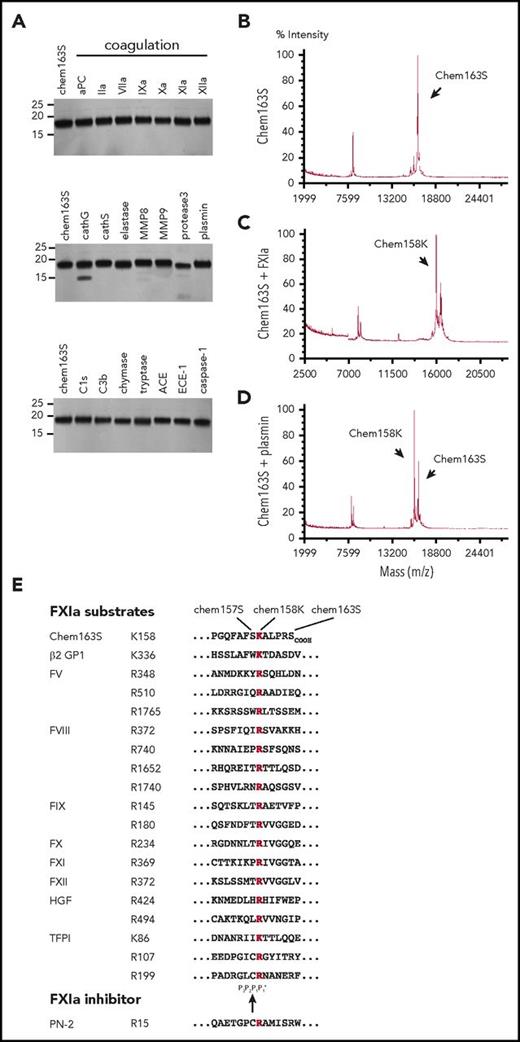
We screened a panel of 22 enzymes involved in the coagulation, fibrinolysis, and inflammation cascades to identify those able to cleave prochemerin by incubating them with purified chem163S and analyzing the cleavage products by MALDI-TOF mass spectrometry (Table 1). None generated active chemerin, chem157S, and only FXIa, plasmin, kallikrein, and tryptase generated detectable chem158K. The level of chem158K formed by treatment with kallikrein was minimal compared with FXIa (supplemental Figure 1). There was a minor reduction in cleavage when HMWK was included with FXIa. SDS-PAGE and MALDI-TOF analysis showed that elastase, MMP8, protease 3, and cathepsin G also generated smaller bands from cleavage at other sites and were not studied further (Figure 1A). Thus, FXIa is the only protease in the coagulation cascade that cleaves chem163S (Figure 1B) specifically into chem158K at a significant rate (Figure 1C), similar to plasmin (Figure 1D). Chemerin and other FXIa substrates are listed in Figure 1E with the conserved basic amino acid at the cleavage site with the residue present in the chem163S P2, P3 and P'1 sites being similar to those in other FXIa substrates.
Mass spectrometric analysis of prochemerin 163S cleavage by FXIa, plasmin, and other proteases
| . | Chemerin forms, molecular weight, Da . | |||||
|---|---|---|---|---|---|---|
| 154F . | 155A . | 156F . | 157S . | 158K . | 163S . | |
| Expected molecular weight | 15571.8 | 15642.8 | 15790.0 | 15877.1 | 16005.3 | 16529.9 |
| FXIa | 16019.4 | |||||
| Plasmin | 16003.7 | |||||
| Cathepsin G | 15792.8 | |||||
| Chymase | 15570.0 | 15785.8 | ||||
| Elastase | 15647.3 | 15881.7 | ||||
| Protease 3 | 15648.0 | |||||
| Tryptase | 16011.4 | |||||
| MMP-8 | 15640.4 | |||||
| . | Chemerin forms, molecular weight, Da . | |||||
|---|---|---|---|---|---|---|
| 154F . | 155A . | 156F . | 157S . | 158K . | 163S . | |
| Expected molecular weight | 15571.8 | 15642.8 | 15790.0 | 15877.1 | 16005.3 | 16529.9 |
| FXIa | 16019.4 | |||||
| Plasmin | 16003.7 | |||||
| Cathepsin G | 15792.8 | |||||
| Chymase | 15570.0 | 15785.8 | ||||
| Elastase | 15647.3 | 15881.7 | ||||
| Protease 3 | 15648.0 | |||||
| Tryptase | 16011.4 | |||||
| MMP-8 | 15640.4 | |||||
A total of 10 μM chem163S was incubated for 30 minutes with 100 nM protease before analysis by mass spectrometry. The molecular weight of the observed products was compared with the expected molecular mass.
Screening of enzymes for cleavage of chem163S. (A) Separation of cleavage products of 10 μM chem163S after 30 minutes of incubation with 100 nM of the labeled proteases by SDS-PAGE followed by silver staining. Molecular weight markers are shown on the left. (B-D). Mass spectrometric analysis of input protein chem163S (B) and reaction mixtures of FXIa (C) and plasmin (D) described in panel A. (E) FXIa cleavage sequences. Alignments extracted from the MEROPS database56 show the cleavage sites for FXIa marked within the chemerin C-terminal sequence with chemerin forms indicated (chem163S), β-2 glycoprotein 1 (β-2 GP),57 FX, FXI, and FXII, the 2 FXIa cleavage sites in FIX and hepatocyte growth factor (HGF),57,58 the 3 FXIa cleavage sites in FV and tissue factor pathway inhibitor (TFPI),59 the 4 FXIa cleavage sites in FVIII, and the binding site from the FXIa inhibitor, protease nexin-2 (PN-2).60 The basic amino acid at P1 is red and the black arrow indicates the FXIa cleavage site.
Screening of enzymes for cleavage of chem163S. (A) Separation of cleavage products of 10 μM chem163S after 30 minutes of incubation with 100 nM of the labeled proteases by SDS-PAGE followed by silver staining. Molecular weight markers are shown on the left. (B-D). Mass spectrometric analysis of input protein chem163S (B) and reaction mixtures of FXIa (C) and plasmin (D) described in panel A. (E) FXIa cleavage sequences. Alignments extracted from the MEROPS database56 show the cleavage sites for FXIa marked within the chemerin C-terminal sequence with chemerin forms indicated (chem163S), β-2 glycoprotein 1 (β-2 GP),57 FX, FXI, and FXII, the 2 FXIa cleavage sites in FIX and hepatocyte growth factor (HGF),57,58 the 3 FXIa cleavage sites in FV and tissue factor pathway inhibitor (TFPI),59 the 4 FXIa cleavage sites in FVIII, and the binding site from the FXIa inhibitor, protease nexin-2 (PN-2).60 The basic amino acid at P1 is red and the black arrow indicates the FXIa cleavage site.
FXIa cleavage of chemerin
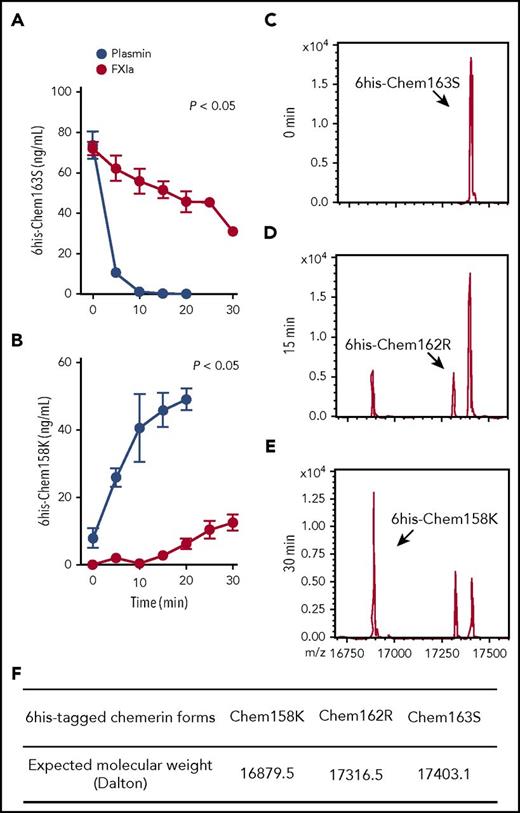
His-tagged chem163S (6his-chem163S) allowed exogenous chem163S to be distinguished from endogenous chem163 and its preexisting products.29,31 6His-chem163S was incubated with FXIa, and the products were analyzed by ELISAs specific for chem163S and chem158K in >80% of 6his-chem163S within 5 minutes, with a corresponding rapid rise in chem158K, and the cleavage was essentially complete in 10 minutes (Figure 2A-B). In contrast, FXIa cleavage of prochemerin had a slower time course, with 22% cleavage of 6his-chem163S by 10 minutes of incubation (Figure 2A). The generation of chem158K by FXIa was not detectable until after 15 minutes of incubation. After 30 minutes of incubation, FXIa had generated 35% of the amount of chem158K as plasmin (Figure 2B).
FXIa cleavage of 6his-chem163S produces 6his-chem162R and 6his-chem158K. (A-B) Purified 6his-chem163S was incubated in assay buffer in the presence of 100 nM FXIa or 100 nM plasmin. Aliquots of reactions were removed every 5 minutes for 30 minutes, PPACK and ethylenediaminetetraacetic acid were added and analyzed by 6his-chem163S ELISA (A) or 6his-chem158K ELISA (B). Data points are the mean ± standard error of the mean from 3 independent experiments. (C-E) Purified 6his-chem163S was incubated with 30 nM FXIa for 0 minutes (C), 15 minutes (D), and 30 minutes (E) and analyzed by MALDI-TOF mass spectrometry. A representative experiment is shown. (F) The expected molecular weights of 6his-tagged chemerin forms were summarized to identify corresponding molecules in panels C-E.
FXIa cleavage of 6his-chem163S produces 6his-chem162R and 6his-chem158K. (A-B) Purified 6his-chem163S was incubated in assay buffer in the presence of 100 nM FXIa or 100 nM plasmin. Aliquots of reactions were removed every 5 minutes for 30 minutes, PPACK and ethylenediaminetetraacetic acid were added and analyzed by 6his-chem163S ELISA (A) or 6his-chem158K ELISA (B). Data points are the mean ± standard error of the mean from 3 independent experiments. (C-E) Purified 6his-chem163S was incubated with 30 nM FXIa for 0 minutes (C), 15 minutes (D), and 30 minutes (E) and analyzed by MALDI-TOF mass spectrometry. A representative experiment is shown. (F) The expected molecular weights of 6his-tagged chemerin forms were summarized to identify corresponding molecules in panels C-E.
MALDI-TOF analysis of the reaction products after 15 minutes revealed that FXIa cleaved 6his-chem163S (Figure 2C) into 2 different species, 6his-chem158K and 6his-chem162R, a previously unreported chemerin form (Figure 2D,F). Incubation with FXIa for 30 minutes resulted in more 6his-chem158K and slightly more 6his-chem162R, but no other chemerin forms were detected (Figure 2E). These results confirm the ELISA data showing that FXIa can cleave chem163S into chem158K and reveal the existence of chem162R.
Cleavage of chem163S at Arg162 is not necessary for generation of chem158K
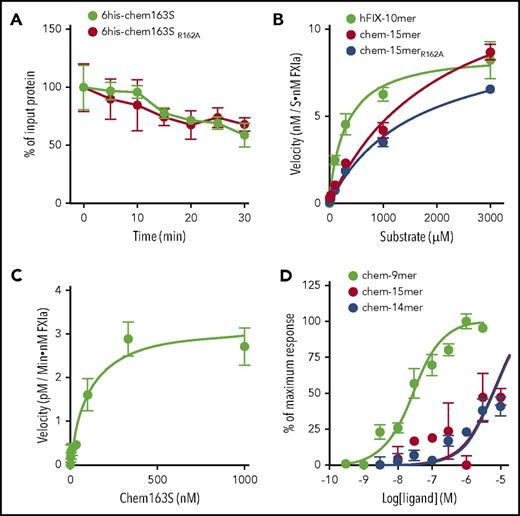
To determine if FXIa cleavage of chemerin at Arg162 is a necessary step in the formation of chem158K, we produced a mutant his-tagged chem163S with Arg162 substituted with Ala (6his-chem163SR162A) and compared FXIa cleavage of 6his-chem163S and 6his-chem163SR162A. There is no difference in substrate consumption rate by FXIa between the wild-type and mutant chemerin proteins (Figure 3A), indicating that cleavage at Arg162 was not necessary for generation of chem158K. This was confirmed by FXIa cleavage of a mutant C-terminal chem-15mer peptide, with Ala substituted for Arg162 (chem-15merR162A), demonstrating a similar catalytic efficiency to wild-type chem-15mer (Figure 3B, Table 2).
Arg162is inactive and not an obligate intermediate necessary for the generation of chem158K by FXIa. (A) Ten mM 6his-chem163S and 10 nM 6his-chem163SR162A were incubated in assay buffer in the presence of 30 nM FXIa. Aliquots of reactions were removed and stopped at the indicated time and analyzed by 6his-chem163S ELISA. Each experiment was repeated ≥3 times. (B) FIX-10mer, chem-15mer, and chem-15merR162A were incubated with 30 nM FXIa for 30 minutes before analysis by HPLC. The product concentration was calculated by interpolation from a standard curve, and the velocities of product generation were determined. (C) Chem163S full-length protein was incubated with 30 nM FXIa for 30 minutes, and products were analyzed by ELISA. (D) Ca++ flux in hCMKLR1/L1.2 cells was followed after the addition of the peptides chem-9mer, chem-14mer and chem-15mer, representing the C-terminal sequence of chem157S, chem162R, and chem163S, respectively. The maximum fluorescence intensities triggered by the addition of each peptide were plotted and used to determine the EC50 value. Data from 4 independent experiments were pooled, and the data are presented as the mean ± standard error of the mean.
Arg162is inactive and not an obligate intermediate necessary for the generation of chem158K by FXIa. (A) Ten mM 6his-chem163S and 10 nM 6his-chem163SR162A were incubated in assay buffer in the presence of 30 nM FXIa. Aliquots of reactions were removed and stopped at the indicated time and analyzed by 6his-chem163S ELISA. Each experiment was repeated ≥3 times. (B) FIX-10mer, chem-15mer, and chem-15merR162A were incubated with 30 nM FXIa for 30 minutes before analysis by HPLC. The product concentration was calculated by interpolation from a standard curve, and the velocities of product generation were determined. (C) Chem163S full-length protein was incubated with 30 nM FXIa for 30 minutes, and products were analyzed by ELISA. (D) Ca++ flux in hCMKLR1/L1.2 cells was followed after the addition of the peptides chem-9mer, chem-14mer and chem-15mer, representing the C-terminal sequence of chem157S, chem162R, and chem163S, respectively. The maximum fluorescence intensities triggered by the addition of each peptide were plotted and used to determine the EC50 value. Data from 4 independent experiments were pooled, and the data are presented as the mean ± standard error of the mean.
Hydrolysis of chemerin peptides, FIX peptides, and chemerin full-length protein by FXIa
| Substrate . | Km, μM . | kcat, min–1 . | kcat/Km, M–1 × min–1 . |
|---|---|---|---|
| FIX-10mer | 300 ± 64 | 5.0 ± 0.2 | 1.7 × 104 |
| chem-15mer | 2120 ± 524 | 8.1 ± 1.3 | 3.8 × 103 |
| chem-15merR162A | 1597 ± 230 | 5.5 ± 0.5 | 3.4 × 103 |
| Chem163S | 103 ± 35 | 54.6 ± 5.3 | 5.3 × 108 |
| Substrate . | Km, μM . | kcat, min–1 . | kcat/Km, M–1 × min–1 . |
|---|---|---|---|
| FIX-10mer | 300 ± 64 | 5.0 ± 0.2 | 1.7 × 104 |
| chem-15mer | 2120 ± 524 | 8.1 ± 1.3 | 3.8 × 103 |
| chem-15merR162A | 1597 ± 230 | 5.5 ± 0.5 | 3.4 × 103 |
| Chem163S | 103 ± 35 | 54.6 ± 5.3 | 5.3 × 108 |
Substrates were digested with FXIa as described in “Materials and methods.” The values for Km, kcat, and kcat/Km were compared between peptides derived from chemerin and FIX, as well as chemerin full-length protein. Data are mean ± SEM; n = 3.
Kinetics of FXIa cleavage of chem163S peptides and protein
We studied the cleavage kinetics of chem163S peptides and full-length protein by FXIa (Table 2). In peptide cleavage assays, the cleavage rate of chem163S peptides was compared with that of the FIX peptide representing the FXIa cleavage sites.32 Chem163S peptides consisting of the C-terminal 15 amino acids of prochemerin (chem-15mer) were subjected to FXIa cleavage followed by HPLC analysis, using FIX peptides as the positive control (Figure 3B). Hydrolysis of chem-15mer by FXIa showed a Km value of 2120 ± 524 μM, a kcat value of 8.1 ± 1.3 min−1, and kcat/Km of 3.8 ± 103 M−1 × min−1. In comparison, cleavage of the FIX peptide by FXIa gave a Km value of 300 ± 64 μM, a kcat value of 5.0 ± 0.2 min−1, and a kcat/Km value of 1.7 ± 104 M−1 × min−1. Thus, the catalytic efficiency (kcat/Km) of the FXIa cleavage of the FIX peptide was approximately threefold more efficient than chemerin peptide cleavage, with the difference mainly due to a decrease in Km. FXIa cleavage of Chem163S full-length protein gave a Km value of 103 ± 35 nM, a kcat value of 54.6 ± 5.3 min−1, and a kcat/Km value of 5.3 ± 108 M−1 × min−1 (Figure 3C), very similar to FXIa cleavage of FIX protein (Km of 53 ± 8 nM, kcat of 33 ± 1 min−1).33
Chem162R has minimal biological activity
We compared the activity of the newly identified chemerin form, chem162R, to other forms of chemerin using their C-terminal peptides to measure Ca++ flux in hCMKLR1/L1.2 cells (Figure 3D). The 50% effective concentration (EC50) for the chem162R peptide was 6.0 ± 1.2 μM, which is comparable to the EC50 of prochemerin chem163S peptide (5.4 ± 1.4 μM), whereas the most active form, chem157S peptide, was ∼150-fold more potent with an EC50 of 28.1 ± 11.6 nM. Thus, the generation of chem162R from chem163S by FXIa did not lead to an increase in biological activity.
FXIa cleavage of chemerin in plasma
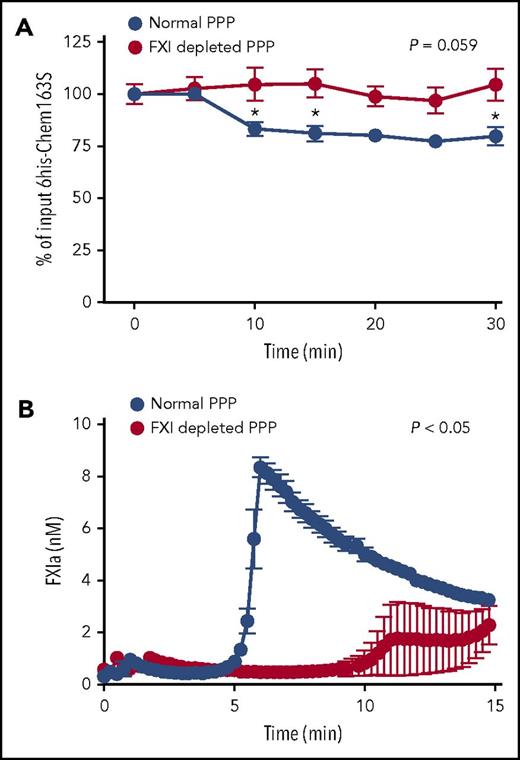
To investigate if FXIa cleaves chem163S in a biological milieu, we triggered the contact pathway with kaolin to activate FXI in human plasma and monitored the consumption of added 6his-chem163S by the 6his-chem163S ELISA. There was an ∼25% decrease of 6his-chem163S after 30 minutes, with most cleavage occurring 5 to 10 minutes after activation (Figure 4A).
6his-chem163S was cleaved in control plasma with contact pathway activation, but not in FXI-depleted plasma. The contact pathway was initiated by the addition of kaolin to control or FXI-depleted, plasma-containing microparticles, Ca++, and GPRP. (A) 6His-chem163S (10 μM) was incubated in control plasma and FXI-depleted plasma with contact pathway activation. Aliquots of reactions were removed every 5 minutes, and the level of 6his-chem163S in each aliquot was determined by 6his-chem163S ELISA. (B) FXIa fluorogenic substrate D-LPR-ANSNH-C3H7•2HCl was incubated in control and FXI-depleted plasma with contact pathway activation. The signal was monitored every 15 seconds, and FXIa concentrations were calculated from the interpolation of a FXIa standard curve. Data from 4 independent experiments were pooled to show mean ± standard error of the mean.
6his-chem163S was cleaved in control plasma with contact pathway activation, but not in FXI-depleted plasma. The contact pathway was initiated by the addition of kaolin to control or FXI-depleted, plasma-containing microparticles, Ca++, and GPRP. (A) 6His-chem163S (10 μM) was incubated in control plasma and FXI-depleted plasma with contact pathway activation. Aliquots of reactions were removed every 5 minutes, and the level of 6his-chem163S in each aliquot was determined by 6his-chem163S ELISA. (B) FXIa fluorogenic substrate D-LPR-ANSNH-C3H7•2HCl was incubated in control and FXI-depleted plasma with contact pathway activation. The signal was monitored every 15 seconds, and FXIa concentrations were calculated from the interpolation of a FXIa standard curve. Data from 4 independent experiments were pooled to show mean ± standard error of the mean.
To demonstrate that the cleavage of chem163S in plasma was due to FXIa, we compared 6his-chem163S cleavage in plasma that had been immune depleted of FXI (to a level of <1% normal FXI) with control plasma. There was no decrease in 6his-chem163S in FXI-depleted plasma (Figure 4A). The inclusion of a fluorogenic substrate specific for FXIa activity demonstrated the generation of peak FXIa activity by 6 minutes in control plasma, followed by a gradual reduction in its activity (Figure 4B). In FXI-depleted plasma, FXIa activity was barely detectable (Figure 4B). The time of maximum FXIa activity in normal plasma correlates with maximal consumption of 6his-chem163S, with its subsequent inhibition probably accounting for the lack of further cleavage after 10 minutes.
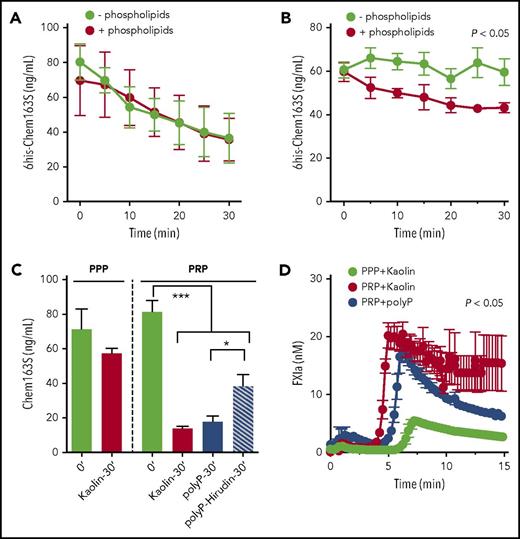
Phospholipids and platelets enhance FXIa cleavage of chem163S in plasma
Because phospholipids and polyP are cofactors for the activation of coagulation pathways,34-38 we determined the effect of phospholipids and polyP on FXIa cleavage of chemerin. PolyP did not modify the rate of FXIa cleavage of chem163S in a purified system (data not shown). Phospholipids did not enhance the cleavage of chem163S by FXIa in assay buffer (Figure 5A), whereas they significantly enhanced FXIa cleavage of chemerin in plasma (Figure 5B). Because the enhancement of cleavage of chem163S by phospholipids suggests a role for lipid surfaces in either chem163S cleavage or increasing FXIa levels, we performed cleavage of endogenous chemerin in PPP supplemented with phospholipids and PRP. The cleavage of endogenous chem163S was modest in PPP supplemented with phospholipids (Figure 5C). With the presence of platelets, there was an 82.7% decrease in endogenous chem163S after kaolin activation of the contact pathway and a 78% decrease in chem163S after polyP activation (Figure 5C). By adding the thrombin inhibitor, hirudin, which blocks the positive feedback of thrombin activation of FXI, cleavage of chem163S was reduced to 53.2% (Figure 5C). The cleavage of chem163S in PRP after contact pathway activation by either kaolin or polyP correlated with the maximum FXIa activity at 5 minutes (Figure 5D).
Microparticles and platelets enhance FXI activation in plasma to cleave chem163S. (A) Purified 6his-chem163S was incubated with FXIa in assay buffer with or without microparticles. Aliquots were removed every 5 minutes for 30 minutes, and the level of 6his-chem163S was measured with the 6his-chem163S ELISA. (B-D) Contact pathway was initiated by kaolin or Polyp in PRP and PPP. (B) Purified 6his-chem163S (10 μM) was incubated in control plasma with or without phospholipids after contact pathway activation. Aliquots of reactions were removed every 5 minutes for 30 minutes, and the levels of 6his-chem163S in each aliquot were analyzed by 6his-chem163S ELISA. (C) After activation by kaolin or Polyp for 30 minutes, the concentration of endogenous chem163S was determined by the chem163S ELISA. In some experiments, hirudin (1.5 U/mL) was added to block thrombin activity. (D) FXIa fluorogenic substrate D-LPR-ANSNH-C3H7•2HCl was incubated in PPP or PRP after contact pathway activation. Fluorescence was monitored every 15 seconds, and the FXIa concentration was calculated by interpolation from a FXIa standard curve. Data from 3 independent experiments were pooled to show mean ± standard error of the mean.
Microparticles and platelets enhance FXI activation in plasma to cleave chem163S. (A) Purified 6his-chem163S was incubated with FXIa in assay buffer with or without microparticles. Aliquots were removed every 5 minutes for 30 minutes, and the level of 6his-chem163S was measured with the 6his-chem163S ELISA. (B-D) Contact pathway was initiated by kaolin or Polyp in PRP and PPP. (B) Purified 6his-chem163S (10 μM) was incubated in control plasma with or without phospholipids after contact pathway activation. Aliquots of reactions were removed every 5 minutes for 30 minutes, and the levels of 6his-chem163S in each aliquot were analyzed by 6his-chem163S ELISA. (C) After activation by kaolin or Polyp for 30 minutes, the concentration of endogenous chem163S was determined by the chem163S ELISA. In some experiments, hirudin (1.5 U/mL) was added to block thrombin activity. (D) FXIa fluorogenic substrate D-LPR-ANSNH-C3H7•2HCl was incubated in PPP or PRP after contact pathway activation. Fluorescence was monitored every 15 seconds, and the FXIa concentration was calculated by interpolation from a FXIa standard curve. Data from 3 independent experiments were pooled to show mean ± standard error of the mean.
Changes in levels of endogenous chemerin forms in PPP and PRP after contact phase activation
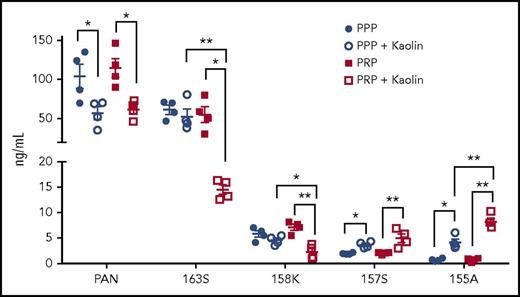
To examine the levels of the different endogenous chemerin forms after activation of the contact pathway, we compared their levels in PPP and PRP by our panel of specific ELISAs (Figure 6). Before contact phase activation, there were no differences in the levels of total chemerin, chem163S, chem158K, chem157S, and chem155A between PPP and PRP. On contact phase activation, there was a significant decrease in both chem163S and chem158K levels in PRP (n = 4; P < .01), with no detectable changes in these levels in PPP, indicating that platelets were critical for FXI activation and subsequent chem163S cleavage. There was a reduction of chemerin detected by the panchemerin ELISA, probably due to the generation of chemerin forms too small to be detected in any of these ELISAs. The concentration of chem157S and chem155A increased in both PPP and PRP after contact phase activation (P < .05), with a greater increase observed in PRP (P < .01). Thus, FXI activation and subsequent cleavage of chem163S is greatly enhanced by the presence of platelets and suggests that processing of chem158K is increased in activated PRP.
Levels of chemerin isoforms before and after contact phase activation in PPP and PRP. Total chemerin (PAN), chem163S, chem158K, chem157S, and chem155A levels in PPP and PRP from donors (n = 4) were determined using specific ELISAs as described under “Experimental procedures.” Horizontal lines show the mean. *P < .05; **P < .01.
Levels of chemerin isoforms before and after contact phase activation in PPP and PRP. Total chemerin (PAN), chem163S, chem158K, chem157S, and chem155A levels in PPP and PRP from donors (n = 4) were determined using specific ELISAs as described under “Experimental procedures.” Horizontal lines show the mean. *P < .05; **P < .01.
Quantification of chemerin forms in plasma from FXI-deficient patients after contact phase activation
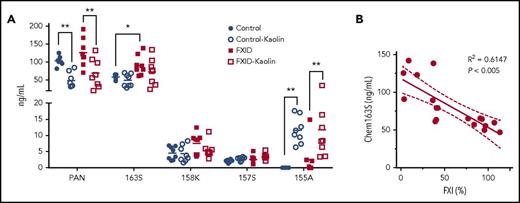
We tested plasma that had been immune-depleted of FII, FVII, FVIII, FIX, FX, FXI, or FXII for cleavage of chem163S on kaolin activation and found that only FXI immune-depleted plasma did not lead to consumption of endogenous chem163S (data not shown). Based on these data, we hypothesized that FXI-deficient patients would have less cleavage of chem163S due to their reduced ability to generate FXIa. To test this, we compared the levels of chemerin forms in PPP from FXI-deficient patients and matched controls using our panel of specific ELISAs (Figure 7A). The chem163S concentration in plasma was significantly higher in FXI-deficient patients (91 ± 10 ng/mL) compared with matched controls (58 ± 3 ng/mL, n = 8; P < .01), whereas the level of FXI was 32% ± 5% in FXI-deficient patients and 95% ± 4% (P < .005) in the control group (Table 3). The levels of FXI in plasma were inversely correlated with the plasma chem163S concentrations (Figure 7B; R2 = 0.6147; P < .005). The total chemerin, chem158K, chem157S, and chem155A ELISA results were similar in the FXI-deficient and control groups. The levels of chem155A were significantly increased in both groups after contact phase activation (P < .05), whereas no changes in chem163S, chem158K, and chem157S levels were detectable.
Levels of chemerin isoforms before and after contact phase activation in controls and FXI-deficient (FXID) patients. (A) Total chemerin (PAN), chem163S, chem158K, chem157S, and chem155A levels in PPP from patients (n = 8) and matching controls (n = 8) were determined using specific ELISAs as described under “Experimental procedures.” Horizontal lines show the mean. *P < .05; **P < .01. (B) The correlation between % FXI levels in plasma and the level of chem163S in plasma was compared (n = 18). Pearson’s correlation coefficient R2 and the P value are shown.
Levels of chemerin isoforms before and after contact phase activation in controls and FXI-deficient (FXID) patients. (A) Total chemerin (PAN), chem163S, chem158K, chem157S, and chem155A levels in PPP from patients (n = 8) and matching controls (n = 8) were determined using specific ELISAs as described under “Experimental procedures.” Horizontal lines show the mean. *P < .05; **P < .01. (B) The correlation between % FXI levels in plasma and the level of chem163S in plasma was compared (n = 18). Pearson’s correlation coefficient R2 and the P value are shown.
Parameters from FXI-deficient patients and matching controls
| . | Control . | FXID . | P . |
|---|---|---|---|
| Chem163S, ng/mL | 58 ± 3 | 91 ± 10 | <.01 |
| FXI, % | 95 ± 4 | 32 ± 5 | <.01 |
| BMI | 22 ± 1 | 25 ± 3 | n.s. |
| Age, y | 44 ± 7 | 45 ± 8 | n.s. |
| Male/female | 5/3 | 5/3 | n.s. |
| . | Control . | FXID . | P . |
|---|---|---|---|
| Chem163S, ng/mL | 58 ± 3 | 91 ± 10 | <.01 |
| FXI, % | 95 ± 4 | 32 ± 5 | <.01 |
| BMI | 22 ± 1 | 25 ± 3 | n.s. |
| Age, y | 44 ± 7 | 45 ± 8 | n.s. |
| Male/female | 5/3 | 5/3 | n.s. |
Data are mean ± SEM; n = 8.
n.s., not significant.
We next examined fresh PRP from 2 FXI-deficient patients and activated contact pathway using polyP (Table 4). Chem163S concentrations were 152 ng/mL and 165 ng/mL in these FXI-deficient PRP. After polyP activation, prochemerin decreased to 48 ng/mL (32% of input prochemerin) and 94 ng/mL (57% of input prochemerin) without FXI repletion and to 29 ng/mL (19% of input prochemerin) and 34 ng/mL (21% of input prochemerin) with FXI repletion. In the control PRP, the baseline chem163S concentrations were 73 ng/mL and 95 ng/mL, which are both lower than those of FXI-deficient PRP, and with polyP activation, the chem163 concentrations were 19 ng/mL (26%) and 12 ng/mL (13%).
Chem163S levels in FXID and control PRP samples from the Stanford Blood Bank
| . | ng/mL . | ng/mL . | ng/mL . |
|---|---|---|---|
| Patient I | 152 | 48 | 29 |
| Patient II | 165 | 94 | 34 |
| Control I | 73 | 19 | N.D. |
| Control II | 95 | 12 | N.D. |
| Polyphosphate | − | + | + |
| Platelets | + | + | + |
| Repleted FXI | − | − | + |
| . | ng/mL . | ng/mL . | ng/mL . |
|---|---|---|---|
| Patient I | 152 | 48 | 29 |
| Patient II | 165 | 94 | 34 |
| Control I | 73 | 19 | N.D. |
| Control II | 95 | 12 | N.D. |
| Polyphosphate | − | + | + |
| Platelets | + | + | + |
| Repleted FXI | − | − | + |
FXID samples are from patients P010 and P011 in supplemental Table 3. PRP samples were treated as described in “Materials and methods.” Chem163S levels were accessed using ELISA. Each value is the average of duplicate readings.
N.D., not done; +, reagent included in assay; −, reagent not included in assay.
Discussion
Although several proteases cleave chem163S, causing its activation and subsequent inactivation,39,40 we carried out a more extensive screening of candidate proteases from the coagulation, fibrinolytic, and inflammatory cascades to identify which were responsible for cleaving chemerin in plasma. We identified FXIa as a protease that cleaves chem163S into chem158K. Comparing known FXIa substrates with the C-terminus of chemerin, there is limited conservation around the cleavage site except the conserved basic amino acid at the P1 position and a preference for an aromatic or hydrophobic amino acid at P3 similar to that observed with peptide substrates.41
Proteases that can generate chem158K are of interest because chem158K is readily converted to chem157S, the most potent form of chemerin, by the plasma carboxypeptidases CPN and CPB2.31 Although chemerin primarily circulates as inactive chem163S in the blood, chem158K is the dominant chemerin form in synovial fluid of patients with inflammatory arthritis and in the cerebral spinal fluid of patients with malignant brain tumors.29 The cleavage rate of chem163S by FXIa is similar to that of FIX, suggesting that chem163S is likely a physiological substrate of FXIa.32,33,42 The Km value for activation of chemerin peptides is ∼20 000-fold higher than the Km value for chemerin full-length protein, likely due to the peptide not being presented in the correct conformation in the absence of the remainder of the protein or to the absence of exosite interactions in substrate recognition.
When we analyzed the cleavage products of chem163S by FXIa, we identified a new chemerin form, chem162R, by mass spectrometry, which could be further cleaved into chem158K. By alanine mutation in both protein and peptide assays, the cleavage at Arg162 in chem163S was not essential for the generation of chem158K, whereas the activity of chem162R was similar to that of chem163S and much less active than chem157S. Thus, chem162R is probably a transient cleavage intermediate with no intrinsic biological role.
Phospholipids enhanced the cleavage of 6his-chem163S in PPP, but did not affect the consumption of 6his-chem163S when incubated with FXIa in buffer, showing that there was no direct effect on the enzymatic activity of FXIa. The extensive cleavage of chem163S after contact phase activation in PRP correlates with the peak generation of FXIa, demonstrating the key role of platelets, which may be increasing the levels of FXIa generation or enhancing chem163S cleavage. Although the precise mechanism(s) of FXI interactions with the platelet surface remains to be fully defined, platelets play a critical role in governing FXI function.43 The enhancement of chem163 cleavage by FXIa in the presence of platelets might be a direct effect on the rate of reaction or an indirect effect, for example, a result of an increase in FXII activation due to polyP secretion.44-46 There was no direct effect of polyP on FXIa cleavage of chem163S in buffer, unlike its enhancement of FXIa cleavage of FV and tissue factor pathway inhibitor.47,48 In FXI-deficient patients, there is no clear correlation between clinical bleeding and plasma FXI coagulant activity, FXI antigen level, activated partial clotting time, or genotype. Recently, thrombin generation measured in PRP, but not PPP, was found to identify bleeding phenotype in FXI deficiency,49 which is consistent with our observation of the key role of platelets in FXI activation and its cleavage of chem163S.
Although chem158K is the final product generated by FXIa in buffer, further processing occurs in plasma. The presence of platelets enhances not only chem163S cleavage, but also the generation of chem157S and chem155A, which could be the result of FXIa-generated chem158K. This indicated that further chemerin cleavage from chem158K in vivo involves other proteases directly or indirectly activated by the contact phase, such as CPB2. CPN and CPB2 can cleave chem158K to chem157S.31 Enzymes released from infiltrating leukocytes were reported to cleave chem157S into chem155A, such as proteinase 3, elastase, and angiotensin-converting enzyme,15,39,50,51 and might represent a physiological step to eliminate local chemerin bioactivity.
In normal plasma, the level of chem158K is low, suggesting that there is some constitutive chemerin processing in vivo. Plasma from healthy individuals also contains detectable activation peptides of FIX, FX, and prothrombin, which is consistent with the notion that the coagulation system has a background turnover.52 Because FXIa can cleave chem163S to generate chem158K, we hypothesized that FXIa is responsible for baseline chem158K generation. To test this, we investigated the levels of chemerin forms in FXI-deficient patient plasma and found significantly higher levels of chem163S than in the matched control group, and the levels of plasma FXI were inversely correlated with plasma chemerin levels, supporting the hypothesis.
This study shows that, in plasma, a contact pathway protease, FXIa, is critical for the formation of active chemerin, a chemoattractant and adipokine. FXI-deficient mice had increased survival and less leukocyte accumulation into the peritoneum in severe polymicrobial peritonitis.9 An elevated serum chemerin level, along with an increased neutrophil count, is an independent risk factor for ischemic stroke and carotid plaque stability.53 At sites of thrombosis and inflammation, chem157S is formed through the generation of chem158K by FXIa, followed by the removal of the C-terminal lysine by CPB2 or CPN. The active chemerin then causes leukocyte infiltration into the thrombus as well as signaling to vascular endothelial cells, smooth muscle cells, preadipocytes, and adipocytes,54,55 thus providing a molecular link between coagulation, inflammation, and innate immunity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and volunteers who contributed blood samples.
This work was supported by grant 1RO1 HL57530 from the National Institutes of Health, National Heart, Lung, and Blood Institute and grant I01BX001959 from the Department of Veterans Affairs (both to L.L.). X.G. was supported by National Institutes of Health, National Heart, Lung, and Blood Institute fellowship training grant 5T32HL120824. L.B. and P.G. were supported by FIRB project grant RBFR12W5V5_004 from the Ministry of University and Scientific Research.
Authorship
Contribution: X.G., Y.Y., L.Z., and L.B. performed the experiments; C.B., L.B., and P.G. arranged the collection of samples from FXI-deficient patients and controls; L.L.L. and J.M. conceived the experimental designs; and X.G., Y.Y., L.Z., L.L.L., and J.M. analyzed the data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John Morser, VA Palo Alto Health Care System, Building 101, A4-131, 3801 Miranda Ave, Palo Alto, CA 94304; e-mail: jmorser@stanford.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal