Key Points
Endogenous TCR knockout increases the expression and functional activity of simultaneously transduced TCR (TCR replacement).
TCR replacement results in superior targeting of hematological malignancies by T cells transduced with a non–HLA-restricted γδ TCR.
Abstract
Adoptive transfer of T cells genetically modified to express a cancer-specific T-cell receptor (TCR) has shown significant therapeutic potential for both hematological and solid tumors. However, a major issue of transducing T cells with a transgenic TCR is the preexisting expression of TCRs in the recipient cells. These endogenous TCRs compete with the transgenic TCR for surface expression and allow mixed dimer formation. Mixed dimers, formed by mispairing between the endogenous and transgenic TCRs, may harbor autoreactive specificities. To circumvent these problems, we designed a system where the endogenous TCR-β is knocked out from the recipient cells using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 (Cas9) technology, simultaneously with transduction with a cancer-reactive receptor of choice. This TCR replacement strategy resulted in markedly increased surface expression of transgenic αβ and γδ TCRs, which in turn translated to a stronger, and more polyfunctional, response of engineered T cells to their target cancer cell lines. Additionally, the TCR-plus-CRISPR–modified T cells were up to a thousandfold more sensitive to antigen than standard TCR-transduced T cells or conventional model proxy systems used for studying TCR activity. Finally, transduction with a pan-cancer–reactive γδ TCR used in conjunction with CRISPR/Cas9 knockout of the endogenous αβ TCR resulted in more efficient redirection of CD4+ and CD8+ T cells against a panel of established blood cancers and primary, patient-derived B-cell acute lymphoblastic leukemia blasts compared with standard TCR transfer. Our results suggest that TCR transfer combined with genome editing could lead to new, improved generations of cancer immunotherapies.
Introduction
Adoptive transfer of genetically engineered T cells has become one of the most promising avenues of cancer immunotherapy. Numerous trials have shown objective clinical responses, and even complete remissions, after adoptive cell transfer in patients with cancers resistant to other therapeutic interventions.1-6 The genetic retargeting of T cells to cancer can be achieved either by transduction with a chimeric antigen receptor (CAR) or a T-cell receptor (TCR) specific for an antigen of choice. Although CAR-based therapy has proven extremely successful in hematological malignancies positive for CD19,7 CARs can only target surface-expressed molecules. In contrast, use of cancer-specific TCRs allows targeting of intracellular proteome and/or metabolome.8
Vertebrate TCRs exist as heterodimers composed of either αβ or γδ TCR chains. Conventional αβ TCRs recognize short antigenic peptides presented by major histocompatibility complex (MHC) I or II molecules (by CD8+ and CD4+ T cells, respectively). The targets recognized by human γδ T cells tend to be predominantly proteins expressed on the cell surface in the context of a generalized cellular stress, including malignant transformation.9 A notable exception to this rule is recognition of pyrophosphate metabolites from the mevalonate pathway (henceforth referred to as phosphoantigens) by the predominant peripheral blood subset of γδ T cells that express TCRs composed of the Vγ9 and Vδ2 chains.10 Because there is no evidence for MHC restriction of γδ T cells, and their targets are expressed on a broad range of cancers, γδ TCRs offer an exciting potential for pan-population immunotherapy.11
The use of a transgenic TCR in primary, patient-autologous T cells is hampered by the presence of preexisting, endogenous TCRs within these cells. Expression of TCRs at the cell surface requires the formation of a ternary complex with the CD3 components of this receptor that constitute a limiting factor for surface expression of the antigen-binding chains of the TCR. As a result, successful expression of transduced TCRs at the cell surface requires that it must successfully compete with the endogenous TCR chains for CD3 association.12 In addition, there is also potential for the formation of hybrid TCRs due to mispairing of endogenous and transduced TCR chains (so-called mixed TCR dimers). Thus, a transduced T cell has potential to express 4 distinct TCRs, only 1 of which is desired. Mixed TCR dimers can also exhibit unpredictable, and potentially dangerous, target specificities, and have been shown to cause fatal autoimmunity.13
Several methodologies have been explored to overcome the issue of TCR competition and mispairing. These approaches include generation of affinity-enhanced TCRs,14 engineering of mutations to improve the pairing of transgenic TCRs,15 or overexpression of CD3 components.12 Affinity-enhanced TCRs have shown high rates of objective clinical response because even a small number of functional TCR molecules is sufficient to convey antigen-specific signaling due to superphysiological activity.16 However, affinity-enhanced, engineered TCRs have bypassed the rigors of thymic selection and have the potential to react to self-antigens. Indeed, unanticipated cross-reactivity by an affinity-enhanced MAGE A3-specific TCR with an epitope from titin caused fatal autoreactivity in both patients who were treated with T cells expressing this TCR.17,18
Here, we aimed to enhance the functionality of natural TCRs during TCR gene transfer of primary CD8+ and CD4+ T cells by simultaneous knockout of the endogenous αβ TCR during transfer of a TCR of choice. This approach enhanced the expression of the transduced TCR at the T-cell surface and resulted in TCR transductants that displayed substantially improved antigen sensitivity. In particular, we focused on leveraging broadly cancer-reactive γδ TCRs in the TCR transfer system as this approach can be used irrespective of patient HLA type. T cells transduced with this system were shown to have superior in vitro and ex vivo reactivity to primary hematological malignancies compared with T cells expressing both endogenous and transgenic TCRs.
Materials and methods
Cell lines and primary cultures
The following cell lines were purchased from ATCC and cultured according to manufacturer’s recommendations: Jurkat E6.1, Molt-3, KBM7, K562, THP-1, U266, TK6. The primary B-cell acute lymphoblastic leukemia (B-ALL) cells (HP, VB, BV, KÖ, CM, PH) were cultured in defined serum-free media as described previously.19,20 The B lymphoblastoid cell line (LCL) 146 was generated by Epstein-Barr virus infection of peripheral mononuclear cells21 (PBMCs) obtained from a healthy donor. Primary B cells and T cells were isolated from PBMCs based on CD19 or CD4 expression, respectively, and used for functional assays 1 day after isolation. The HLA-A2+ melanoma cell line was cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, penicillin/streptomycin, and l-glutamine (all from Gibco, Paisley, United Kingdom). T-cell clone γδ20 was generated by single-cell cloning from PBMCs as described.22 We also made use of an αβ T-cell clone Mel13, specific for a Melan-A epitope EAAGIGILTV presented in the context of HLA-A2.23 T-cell clones and lines were expanded in presence of 1 µg/mL phytohemagglutinin (PHA) and allogeneic irradiated feeders from at least 3 donors.24
Generation of transfer vectors and lentiviral particles
A TCR from clone γδ20 was sequenced in-house using the SMARTer RACE kit (Clontech) and 2-step polymerase chain reaction using universal forward primers and reverse primers specific for constant regions of TCR-γ and TCR-δ. The γδ20 TCR was found to be composed of a Vγ9 and Vδ2 chain. Mel13 is a sister clone of Mel5 and the TCR sequence has been published before.23 We have also produced a TCR-peptide-HLA A2 cocomplex structure of this TCR with analog25 and natural26 antigens. Codon-optimized, full-length TCR chains, separated by a self-cleaving 2A sequence,27 were synthesized (Genewiz) and cloned into the third-generation lentiviral transfer vector pELNS (kindly provided by James Riley, University of Pennsylvania, Philadelphia, PA). The pELNS vector contains a rat CD2 (rCD2) marker gene separated from the TCR by another self-cleaving 2A sequence. For clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein-9 (Cas9) mediated knockout of both TCR-β constant regions (trbc1 and trbc2; International Immunogenetics Information System [IMGT] Web site), 4 guide RNAs (gRNAs) targeting the first exon of the trbc gene segments were designed using, and cloned into, pLentiCRISPR v2 plasmid28 (Addgene plasmid 52961, kindly provided by Feng Zhang, McGovern Institute for Brain Research, Massachusetts Institute of Technology). pLentiCRISPR v2 plasmid encodes SpCas9 protein and a puromycin-resistance marker gene (puromycin N-acetyltransferase [pac]). The sequence alignments of gRNAs are summarized in supplemental Figure 1 (available on the Blood Web site).
Lentiviral particles were generated by calcium chloride transfection of HEK 293T cells. TCR transfer vectors were cotransfected with packaging and envelope plasmids pMD2.G, pRSV-Rev, and pMDLg/pRRE, whereas CRISPR/Cas9 vectors were cotransfected with packaging and envelope plasmids pMD2.G and psPAX2 (all from Addgene). Lentiviral particles were concentrated by ultracentrifugation prior to transduction of T cells.
T-cell transduction
PBMCs were obtained from healthy donors obtained via the Welsh Blood Service. Primary T cells were purified by Ficoll separation followed by magnetic enrichment for either CD8+ or CD4+ T cells (Miltenyi Biotec). T cells were subsequently activated overnight by incubation with CD3/CD28 beads (Dynabeads; Life Technologies) at a 3:1 bead-to-T-cell ratio. After activation, the T cells were transduced with lentiviral particles encoding either a TCR only or both a TCR and CRISPR/Cas9, in the presence of 5 µg/mL polybrene (Santa Cruz Biotechnology). T cells that had taken up the virus were selected by incubation with 2 µg/mL puromycin (Life Technologies) and magnetic enrichment with α-rCD2 phycoerythrin (PE) antibody (clone OX-34, Biolegend) followed by α-PE magnetic beads (Miltenyi Biotec). Fourteen days posttransduction, T cells were expanded with allogeneic feeders.22 For all functional experiments, transduced T cells were >95% rCD2+.
Flow cytometry
For surface staining, 50 000 cells were stained with Fixable Live/Dead Violet Dye (Life Technologies) and the following antibodies: rCD2 fluorescein isothiocyanate (Biolegend), pan-αβ TCR PE, pan-γδ TCR allophycocyanin (APC), and CD4 PE-Vio770 and CD8 APC-Vio770 (where applicable; all obtained from Miltenyi Biotec). Mel13-transduced cells were also stained with a cognate tetramer (HLA-A2 refolded in-house with the EAAGIGILTV epitope) according to the optimized tetramer-staining protocol.29 For characterization of the differentiation phenotype of the transduced T cells, the following antibodies were used: programmed death 1 (PD-1) PE, CCR7 peridinin chlorophyll protein–Vio770, CD45RA PE-Vio770, CD45RO fluorescein isothiocyanate, and CD27 APC (all obtained from Miltenyi Biotec). All cell lines tested were stained with BTN3 PE antibody (Biolegend), with or without zoledronate pretreatment. For the Jurkat activation assay, cells were incubated with antigen for 16 hours and subsequently stained for CD69. For intracellular cytokine staining, T cells were incubated for 5 hours with target cell lines, and stained for CD107a (BD Biosciences), tumor necrosis factor α (TNFα), and interferon γ (IFNγ), according to the manufacturer’s recommendation (all obtained from Miltenyi Biotec). Cells were simultaneously stained for combinations of surface markers rCD2, CD3, CD4, and CD8 as required. Events were acquired on FACSCanto II (BD Biosciences) and analyzed using FlowJo software (TreeStar). Polyfunctionality plots were generated using SPICE software.30 A minimum of 10 000 viable events were collected per sample.
51-Chromium release assay
For the assessment of cytotoxicity, target cells were preincubated with Chromium-51 (Perkin Elmer) and then coincubated with T cells at various effector-to-target (E:T) ratios for 4 hours, as described before.29 Cell lysis was calculated according to the formula: % lysis = (experimental 51Cr release − spontaneous 51Cr release)/(experimental 51Cr release − maximum 51Cr release) × 100%.
Enzyme-linked immunosorbent assay
Briefly, 30 000 T cells were coincubated with 90 000 target cells for 16 hours, and the supernatant was harvested. The concentration of macrophage inflammatory protein 1-β (MIP1-β), TNFα, or IFNγ in supernatant was quantified using the respective detection kit (R&D Systems), according to the manufacturer’s instructions. When indicated, target cells were preincubated with 50 µM zoledronic acid (Sigma Aldrich) for 16 hours and washed extensively before coincubation with T cells.
Data analysis
All data were analyzed in GraphPad Prism software, unless specified otherwise.
Results
Design and validation of simultaneous TCR knockout and transfer (TCR replacement) system
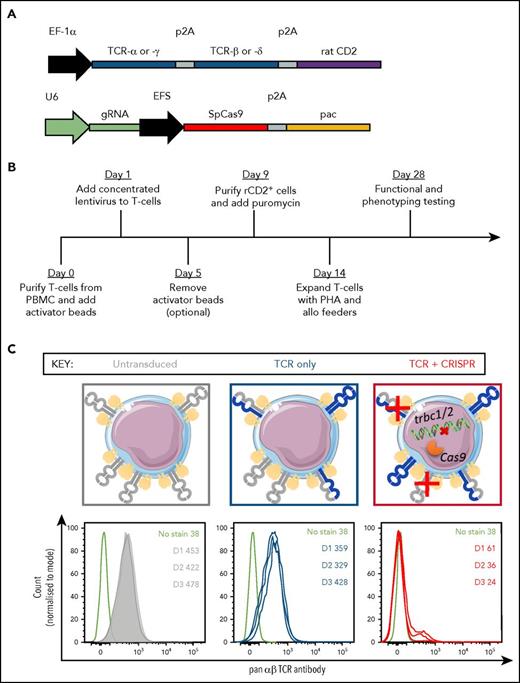
Lentiviral transduction of primary T cells is greatly enhanced when the cells are actively dividing in response to TCR and costimulatory signals.31 To incorporate this enhancement and produce a simple, time-efficient methodology that could be applied with many existing TCR transduction systems, we activated T cells in the presence of 2 separate lentiviral populations, 1 encoding the TCR of choice as a transgene, the other CRISPR/Cas9 targeting the endogenous (but not codon-optimized) TCR-β constant region (trbc1 and trbc2) as described in “Materials and methods.” Four gRNAs targeting TCR-β were designed and showed >90% knockout efficiency in the Jurkat T-cell leukemia line (supplemental Figure 1A). gRNA 1 was selected for use in primary T cells due to the high degree of mismatch between endogenous and the standard, codon-optimized TCR-β sequences generally used during TCR transduction (supplemental Figure 1B). The TCR and CRISPR lentiviruses encoded 2 different selection markers (ectopically expressed rCD2 and puromycin resistance gene, respectively; Figure 1A), allowing selection of cells that had integrated the lentiviral cargo. In addition, the use of rCD2, which was stoichiometrically expressed with the TCR, allowed ready comparison between different donors and different transduction conditions (TCR only or TCR plus CRISPR). Following lentiviral transduction, transduced cells were selected by magnetic or fluorescence-based sorting and culturing with puromycin, where applicable, followed by a conventional T-cell expansion protocol (Figure 1B). Although the selection of transduced cells by rCD2-based purification and puromycin treatment resulted in a nearly 90% decrease in cell number (supplemental Figure 2A), the selected cells were then capable of expanding to the same extent as untransduced cells for at least 5 consecutive expansions with allogeneic feeders and PHA (supplemental Figure 2B). Notably, transduction efficiency with TCR-bearing lentivirus was decreased in the presence of CRISPR lentivirus, indicating that a fraction of cells were capable of accepting only 1 of the lentiviruses (supplemental Figure 2C).
Construct design and validation for transduction of primary T cells. (A) Schematic representation of transgenes cloned into the pELNS vector (top) or lentiCRISPRv2 vector (bottom). (B) Timeline for transduction and selection of primary T cells. (C) Graphical representation of TCR expression on primary T cells transduced with pELNS vector, with and without lentiCRISPRv2 vector (top). Gray molecules represent endogenous TCR chains whereas blue ones represent transduced TCR chains. The histograms below show endogenous αβ TCR expression in 3 donors (gray, untransduced; blue, transduced only with a γδ TCRl red, transduced with a γδ TCR and CRISPR), as well as a representative unstained control (black). The color coding is maintained throughout the manuscript. The numbers on histograms refer to geometric mean fluorescence intensities of αβ TCR expression across 3 donors (D1, D2, D3). EF-1α, elongation factor-1 α promoter; EFS, short EF-1α promoter; pac, puromycin N-acetyltransferase; U6, RNA polymerase III promoter.
Construct design and validation for transduction of primary T cells. (A) Schematic representation of transgenes cloned into the pELNS vector (top) or lentiCRISPRv2 vector (bottom). (B) Timeline for transduction and selection of primary T cells. (C) Graphical representation of TCR expression on primary T cells transduced with pELNS vector, with and without lentiCRISPRv2 vector (top). Gray molecules represent endogenous TCR chains whereas blue ones represent transduced TCR chains. The histograms below show endogenous αβ TCR expression in 3 donors (gray, untransduced; blue, transduced only with a γδ TCRl red, transduced with a γδ TCR and CRISPR), as well as a representative unstained control (black). The color coding is maintained throughout the manuscript. The numbers on histograms refer to geometric mean fluorescence intensities of αβ TCR expression across 3 donors (D1, D2, D3). EF-1α, elongation factor-1 α promoter; EFS, short EF-1α promoter; pac, puromycin N-acetyltransferase; U6, RNA polymerase III promoter.
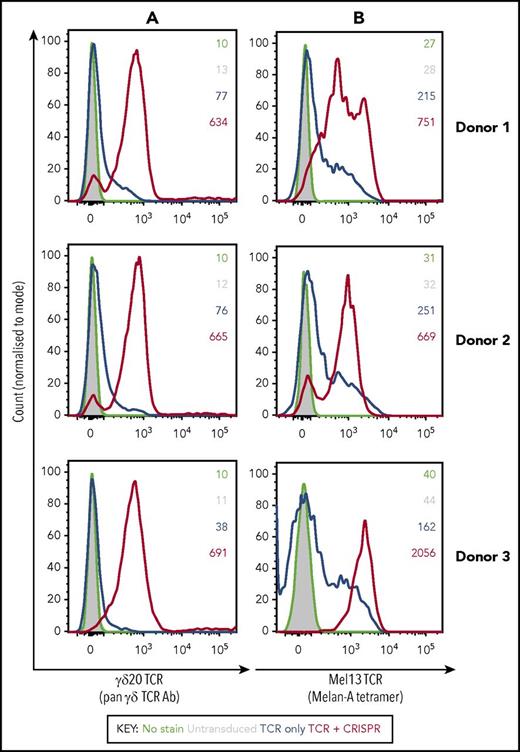
Transduction of primary αβ T cells with a γδ TCR on its own resulted only in a minor downregulation of the endogenous αβ TCR expression. In contrast, αβ TCR expression was almost completely ablated when cells were cotransduced with TCR and CRISPR/Cas9 in all donors tested, showing high efficiency of the TCR replacement system (Figure 1C). We then compared the expression of pyrophosphate metabolite–specific γδ TCR (γδ20), or a melanoma antigen–specific αβ TCR (Mel13), in primary CD8+ αβ T cells that were either single (TCR) or double (TCR plus CRISPR) transduced. Although the expression of transgenic TCRs in single transduced T cells was relatively low (as detected by pan-γδ TCR antibody or a cognate Mel13 tetramer, HLA-A2:EAAGIGILTV), cotransduction with the CRISPR vector resulted in dramatically enhanced expression of the transduced TCR in all donors tested (up to 10-fold increase in mean fluorescence intensity, as well as a distinct shift of histogram peak; Figure 2). A high level of expression of the transgenic αβ TCR in double-transduced cells further confirmed that TCR-β targeting gRNA was unable to cleave the codon-optimized receptor, and that the presence of endogenous TCRα chains did not have a detrimental effect on the expression of the transgenic TCR.
The expression of transduced TCRs in primary CD8+ αβ T cells derived from 3 healthy donor PBMCs is markedly increased in the presence of CRISPR/Cas9 specific for endogenous TCR-β. Histograms represent staining of transduced CD8+ cells with a pan-γδ TCR antibody (A) or with a HLA-A2:EAAGIGILTV tetramer cognate for Mel13 TCR (B), whereas the numbers refer to geometric mean intensity of staining. Black indicates unstained control; gray, untransduced T cells; blue, transduced only with a TCR; red, transduced with a TCR and CRISPR.
The expression of transduced TCRs in primary CD8+ αβ T cells derived from 3 healthy donor PBMCs is markedly increased in the presence of CRISPR/Cas9 specific for endogenous TCR-β. Histograms represent staining of transduced CD8+ cells with a pan-γδ TCR antibody (A) or with a HLA-A2:EAAGIGILTV tetramer cognate for Mel13 TCR (B), whereas the numbers refer to geometric mean intensity of staining. Black indicates unstained control; gray, untransduced T cells; blue, transduced only with a TCR; red, transduced with a TCR and CRISPR.
TCR replacement improves the functional response of transgenic T cells to target cells
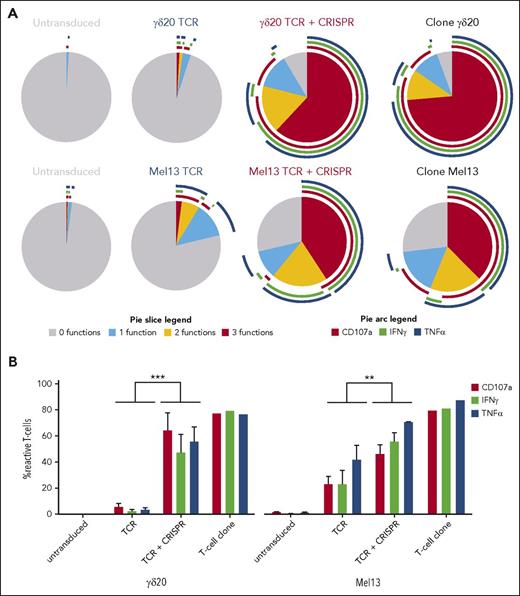
It is generally acknowledged that the number of functional TCR molecules on the surface of a T cell is one of the factors governing T-cell sensitivity to an antigen.32 Therefore, we activated the single- or double-transduced CD8+ T cells with their target cell lines (B-LCL line preincubated with zoledronate for γδ20 TCR, and a HLA-A2+ melanoma cell line for Mel13 TCR), and measured the percentage of cells expressing a marker of cytotoxicity CD107a, and 2 cytokines IFNγ and TNFα. The response of TCR-plus-CRISPR–transduced T cells was markedly stronger than that of cells transduced with only TCR (Figure 3A; supplemental Figure 3). Up to 90% of TCR-plus-CRISPR–transduced cells expressed at least 1 activation marker in response to target cells, and most of these cells expressed all 3 markers tested for, indicating a strong polyfunctional response to antigen. Conversely, <10% and 40% of cells transduced only with γδ20 and Mel13 TCR, respectively, were capable of mounting a response to the target cells, with only a small fraction of the cells that did respond exhibiting >1 function. In comparison, TCR-transduced cells with CRISPR knockout were capable of mounting a statistically significantly stronger response to their cognate antigen in all donors tested (Figure 3B), and the response of TCR-plus-CRISPR cells was comparable to that of parental clones. Both TCR only and TCR-plus-CRISPR cells were capable of downregulating the transgenic TCR upon stimulation with the cognate antigen (supplemental Figure 4). Importantly, the untransduced and single/double-transduced cells showed similar terminally differentiated effector memory phenotype,33 plausibly resulting from CD3/CD28 bead expansion, but no signs of T-cell exhaustion, in terms of PD-1 expression34 (supplemental Figure 5).
The functional response to target cell lines is significantly increased in CD8+ T cells cotransduced with TCR and CRISPR/Cas9 specific for endogenous TCR-β. (A) Polyfunctionality plots representing the response of transduced and untransduced T cells in comparison with the parental T-cell clone. Top row, The response to a B-LCL line preincubated with zoledronate by cells transduced with the γδ20 TCR. Bottom row, Responses to an HLA-A2+ melanoma cell line by cells transduced with the Mel13 αβ TCR. Only viable CD3+ cells were included in the analysis whereas the gates for cells positive for a given function were set based on appropriate fluorescence minus 1 and biological controls. Representative data from 2 independent experiments and 3 donors are shown. (B) The response of transduced T cells to target cell lines, in terms of CD107a, IFNγ, and TNFα expression (mean and standard deviation from 3 donors are shown). The percentage of cells that were positive for a given function in absence of cognate stimulus (ie, T cells plus B-LCL for γδ20 TCR, and T cells alone for Mel13 TCR) was subtracted from the percentage of cells positive in the presence of cognate stimulus (ie, T cells plus B-LCL preincubated with zoledronate or T cells plus HLA-A2+ melanoma cell line, respectively). The statistical significance of difference between the response of cells transduced only with TCR or with TCR plus CRISPR was measured by the paired Student t test. ***P = .0001; **P = .002.
The functional response to target cell lines is significantly increased in CD8+ T cells cotransduced with TCR and CRISPR/Cas9 specific for endogenous TCR-β. (A) Polyfunctionality plots representing the response of transduced and untransduced T cells in comparison with the parental T-cell clone. Top row, The response to a B-LCL line preincubated with zoledronate by cells transduced with the γδ20 TCR. Bottom row, Responses to an HLA-A2+ melanoma cell line by cells transduced with the Mel13 αβ TCR. Only viable CD3+ cells were included in the analysis whereas the gates for cells positive for a given function were set based on appropriate fluorescence minus 1 and biological controls. Representative data from 2 independent experiments and 3 donors are shown. (B) The response of transduced T cells to target cell lines, in terms of CD107a, IFNγ, and TNFα expression (mean and standard deviation from 3 donors are shown). The percentage of cells that were positive for a given function in absence of cognate stimulus (ie, T cells plus B-LCL for γδ20 TCR, and T cells alone for Mel13 TCR) was subtracted from the percentage of cells positive in the presence of cognate stimulus (ie, T cells plus B-LCL preincubated with zoledronate or T cells plus HLA-A2+ melanoma cell line, respectively). The statistical significance of difference between the response of cells transduced only with TCR or with TCR plus CRISPR was measured by the paired Student t test. ***P = .0001; **P = .002.
TCR replacement improves the sensitivity to antigen of a γδ TCR by several orders of magnitude
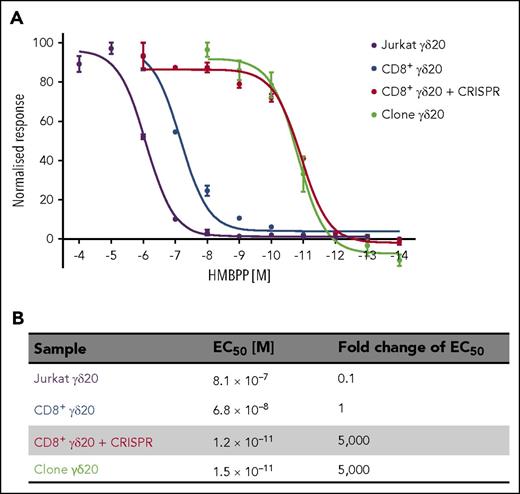
T cells require a given copy number of antigen to be present on target cells in order to mount a successful response, thus defining the antigen sensitivity. Although antigen sensitivity may be manipulated in the case of αβ T cells by affinity maturation of the TCR14 so that it can robustly respond to a very limited number of antigen copies,35,36 no such technology has been developed for γδ TCRs. Therefore, we decided to investigate whether increasing the copy number of γδ TCR on transgenic T cells by CRISPR/Cas9 knockout of endogenous TCRs would increase the sensitivity to the cognate antigen (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP) by TCR-transduced cells. In parallel, we tested the model proxy system for studying the role of TCR in target cell recognition, namely the Jurkat T-cell leukemia line.9 We measured T-cell response to the antigen in terms of MIP-1β secretion (for T cells) or CD69 upregulation (Jurkat). In our experience, MIP-1β secretion has been the most sensitive method for detecting T-cell activation.35-41 Indeed, when we used IFNγ as a readout of T-cell activation, we could not detect any meaningful response from γδ20 TCR-only transduced T cells, whereas the activation of TCR-plus-CRISPR cells closely replicated that of the parental clone (supplemental Figure 6A). Furthermore, the parental T-cell clone was more sensitive to the antigen by 4 orders of magnitude (Figure 4) than the Jurkat cell line (when using MIP-1β and CD69 as markers of activation, respectively). More importantly, TCR-only transduced CD8+ cells were only slightly more sensitive than the Jurkat cell line, and well over a thousandfold less sensitive than the T-cell clone. In contrast, TCR-plus-CRISPR–transduced cells showed a similar degree of antigen sensitivity as the parental clone, and were >50 000 or >5000 more sensitive than Jurkat or single-transduced T cells, respectively. The improvement in antigen sensitivity observed with Mel13 TCR-plus-CRISPR–transduced T cells compared with Mel 13 TCR-only transduced T cells was more modest than for γδ20 TCR (∼10-fold greater sensitivity in terms of MIP-1β production; supplemental Figure 6B-C). Importantly, the improvement observed with Mel13 TCR-plus-CRISPR T cells extended to superior cytotoxic activity against HLA-A2+ melanoma targets compared with cells transduced with Mel13 TCR only (supplemental Figure 7).
The sensitivity to antigen of γδ20 TCR-plus-CRISPR–transduced CD8+ cells is higher by several orders of magnitude than the sensitivity of CD8+ cells transduced only with γδ20 TCR. (A) The sensitivity to the titrated antigen HMBPP was measured either by CD69 mobilization (Jurkat) or MIP-1β production (transduced T cells and T-cell clone) after overnight incubation with the antigen. CD69 mean fluorescence intensity or MIP-1β concentration was normalized by subtracting the values of unstimulated cells, and assuming the maximum value as 100%. The EC50 values were calculated in GraphPad Prism software by nonlinear regression curve fitting. (B) The 50% effective concentration (EC50) represented as molar concentration of antigen and fold change. Representative data of 2 independent experiments carried out in duplicate are shown.
The sensitivity to antigen of γδ20 TCR-plus-CRISPR–transduced CD8+ cells is higher by several orders of magnitude than the sensitivity of CD8+ cells transduced only with γδ20 TCR. (A) The sensitivity to the titrated antigen HMBPP was measured either by CD69 mobilization (Jurkat) or MIP-1β production (transduced T cells and T-cell clone) after overnight incubation with the antigen. CD69 mean fluorescence intensity or MIP-1β concentration was normalized by subtracting the values of unstimulated cells, and assuming the maximum value as 100%. The EC50 values were calculated in GraphPad Prism software by nonlinear regression curve fitting. (B) The 50% effective concentration (EC50) represented as molar concentration of antigen and fold change. Representative data of 2 independent experiments carried out in duplicate are shown.
Endogenous TCR knockout enhances recognition of hematological malignancies via a γδ TCR
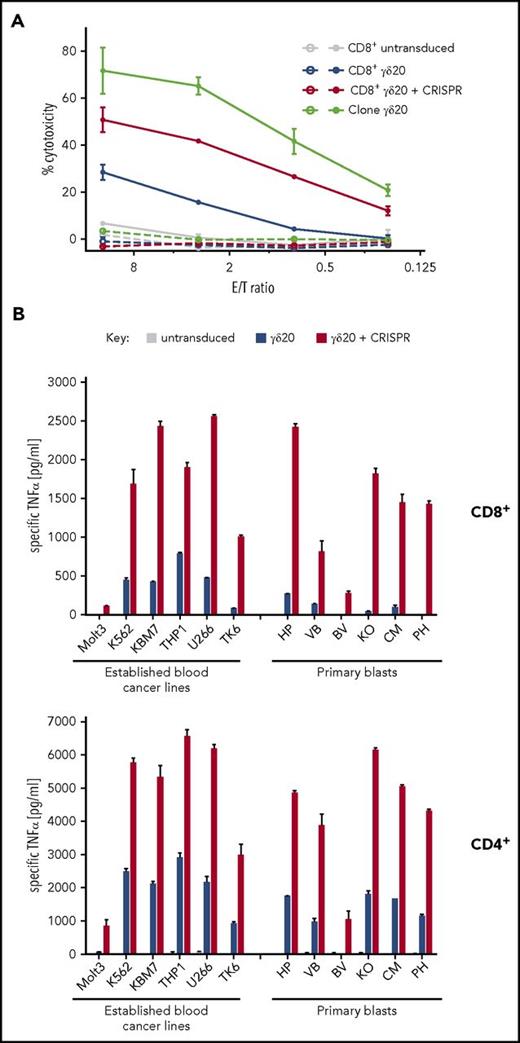
Vγ9Vδ2 TCRs are known to recognize metabolites of the mevalonate pathway in the context of the butyrophilin 3A1 molecule.42-44 The mevalonate pathway is often dysregulated in cancer cells,45 and can be further modulated by aminobisphosphonates such as clinically approved zoledronate.46 Vγ9Vδ2 T cells and TCRs thus have the potential to target multiple different cancer types. Therefore, we first tested the cytotoxic activity of γδ20 TCR-transduced cells against an LCL line derived from the same donor as the parental T-cell clone. In line with the polyfunctionality profile described in Figure 3, TCR-plus-CRISPR–transduced CD8+ cells were able to exhibit stronger cytotoxic activity, especially at low E:T ratios (26% vs 4% at 0.8 E:T; Figure 5A) to LCL preincubated with zoledronate. No cytotoxicity was observed without zoledronate pretreatment, even in the case of the parental T-cell clone, thus indicating that the endogenous accumulation of mevalonate metabolites in that cell line was not sufficient to trigger T-cell activation.
T cells transduced with CRISPR replacement show a markedly stronger response to blood cancer lines than with standard transduction techniques. (A) Four-hour cytotoxicity of transduced CD8+ cells, as well as parental γδ20 T-cell clone, against an untreated (empty symbols) or zoledronate-pretreated (filled symbols) γδ20 donor-autologous B-LCL. Representative data are shown from 3 donors tested in 2 experiments carried out in duplicate. (B) TNFα secretion by transduced CD8+ (top) or CD4+ (bottom) T cells after overnight coincubation with a panel of established blood cancer lines of diverse lymphoid and myeloid origin, or patient-derived B-ALL cells. Cancer cells were preincubated with zoledronate for 24 hours before coincubation with T cells. TNFα secretion was normalized by subtracting TNFα produced by T cells alone, and by cancer cells alone. No specific TNFα secretion by T cells was observed in absence of zoledronate pretreatment. Representative data are shown from 3 donors and 2 experiments carried out in duplicate.
T cells transduced with CRISPR replacement show a markedly stronger response to blood cancer lines than with standard transduction techniques. (A) Four-hour cytotoxicity of transduced CD8+ cells, as well as parental γδ20 T-cell clone, against an untreated (empty symbols) or zoledronate-pretreated (filled symbols) γδ20 donor-autologous B-LCL. Representative data are shown from 3 donors tested in 2 experiments carried out in duplicate. (B) TNFα secretion by transduced CD8+ (top) or CD4+ (bottom) T cells after overnight coincubation with a panel of established blood cancer lines of diverse lymphoid and myeloid origin, or patient-derived B-ALL cells. Cancer cells were preincubated with zoledronate for 24 hours before coincubation with T cells. TNFα secretion was normalized by subtracting TNFα produced by T cells alone, and by cancer cells alone. No specific TNFα secretion by T cells was observed in absence of zoledronate pretreatment. Representative data are shown from 3 donors and 2 experiments carried out in duplicate.
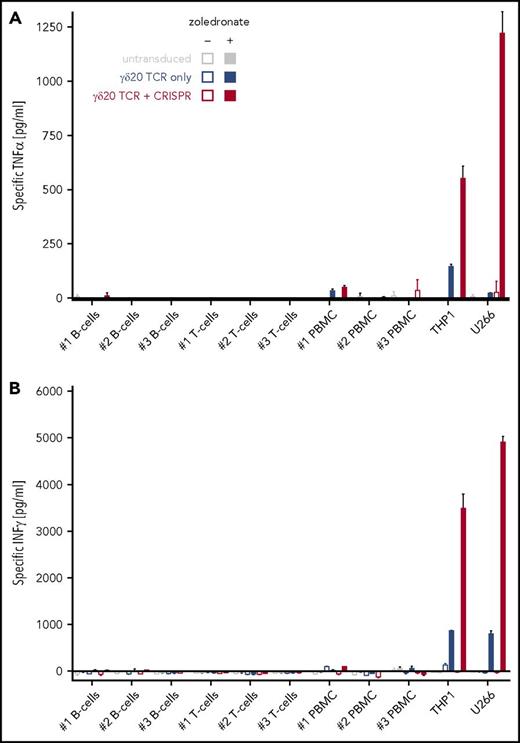
Because Vγ9Vδ2 T cells do not require CD8/CD4 coreceptors for target recognition, we then investigated the potential of the TCR replacement system to redirect both CD8+ and CD4+ T-cell subsets to a panel of hematological malignancies. We tested the ability of single- (γδ20 only) and double-transduced (γδ20 plus CRISPR) T cells to undergo activation and cytokine secretion (TNFα and IFNγ) in response to established blood cancer cell lines (T-cell ALL [T-ALL], acute myeloid leukemia [AML], multiple myeloma) as well as primary, patient-derived B-ALL cells (Figure 5B; supplemental Figure 8). The primary B-ALL cells used here have been previously shown to closely replicate the characteristics of the cancer without cell culture–induced bias.19,20 Single-transduced T cells showed only low reactivity, or no reactivity at all (especially in the case of the Molt3 T-ALL line and primary B-ALL cells), to hematological malignancies pretreated with zoledronate; conversely, TCR-plus-CRISPR–transduced cells responded to all cell lines tested, in a much stronger manner than the TCR-only transduced cells, even to cancer cells expressing an almost undetectable level of BTN3 on the surface (supplemental Figure 9A). No reactivity of γδ20 TCR-transduced cells (with or without TCR-β CRISPR) was observed against freshly isolated, zoledronate-treated healthy cells (Figure 6), despite strong BTN3 expression on the cell surface (supplemental Figure 9B).
Increase of γδ TCR expression by TCR-β knockout does not enhance the targeting of normal cells by engineered T cells. Normal cells were isolated from peripheral blood of 3 healthy donors (PBMC isolation followed by magnetic pullout of CD19+ B cells or CD4+ T cells) and incubated with 50 μM zoledronate (where indicated). On the following day after isolation, the cells were coincubated with transduced T cells for 16 hours, followed by quantification of secreted (A) TNFα or (B) IFNγ. The concentration of secreted cytokines was normalized by subtracting the values from T cells incubated alone and target normal cell incubated alone. Leukemia cell line THP1 and myeloma cell line U266 were included as positive controls. Representative data from 2 TCR-transduced donors are shown.
Increase of γδ TCR expression by TCR-β knockout does not enhance the targeting of normal cells by engineered T cells. Normal cells were isolated from peripheral blood of 3 healthy donors (PBMC isolation followed by magnetic pullout of CD19+ B cells or CD4+ T cells) and incubated with 50 μM zoledronate (where indicated). On the following day after isolation, the cells were coincubated with transduced T cells for 16 hours, followed by quantification of secreted (A) TNFα or (B) IFNγ. The concentration of secreted cytokines was normalized by subtracting the values from T cells incubated alone and target normal cell incubated alone. Leukemia cell line THP1 and myeloma cell line U266 were included as positive controls. Representative data from 2 TCR-transduced donors are shown.
Discussion
TCR gene transfer has been proven as a clinically successful means of redirecting a patient’s immune system to combat different cancer types.47 However, the preexistence of endogenous αβ TCRs in the recipient T cells has limited the clinical use to highly competitive/high-affinity αβ TCRs. Here, we demonstrate that cancer-specific αβ or γδ TCRs that do not compete well with recipient TCRs, and therefore exhibit weak functional activity, can be efficiently used to redirect recipient T cells to cancer if combined with simultaneous knockout of endogenous TCR-β. The resultant engineered T cells were as sensitive to antigen as the starting T-cell clone, suggesting that mispairing between endogenous TCR-α chains and transduced TCR-β must be minimal. This finding is in accordance with the results of Provasi et al using zinc finger nucleases where transgenic TCR activity was comparable in T cells deficient for only the endogenous TCR-β and both TCR-α and -β.48 Furthermore, because TCR-α and -β chains cannot pair with TCR-γ and -δ chains, disruption of just the TCR-β chain is sufficient to achieve the optimal expression of transgenic γδ TCRs.
To date, there have been several reported attempts to combine endogenous TCR knockout, using zinc finger nucleases,48,49 transcription activator-like effector nucleases,50-53 or CRISPR/Cas9,54 with redirecting the T cells to cancer, in most cases via CARs. This is the first report demonstrating successful redirection of primary T cells with a pan-cancer–reactive γδ TCR in combination with endogenous TCR-β knockout. We showed that removal of the endogenous TCR-β chain leads to a striking increase of surface expression of transgenic αβ and γδ TCRs that translates into a much stronger response of engineered T cells to cancer lines. Although it has recently been shown by Eyquem et al that CAR insertion into the TCR locus is beneficial due to limiting and controlling CAR expression by physiological means, thus preventing premature exhaustion, the antigen-binding kinetics and affinity of natural TCRs differ significantly from that of CARs, and therefore a high copy number of TCRs on the cell surface appears more desirable.55 Indeed, CRISPR-plus-TCR–transduced T cells exhibited a significantly more polyfunctional response profile when presented with target cells than that observed with TCR transduction in the absence of TCR-β knockout, without any apparent changes in terms of T-cell differentiation and exhaustion. Importantly, Ding et al showed that polyfunctional T cells are crucial for achieving a successful clinical outcome in patients suffering from hematological malignancies.56 Moreover, this is the first side-by-side comparison of the antigen sensitivity of the model Jurkat T-cell line, primary T cells transduced with a given TCR and the parental T-cell clone using the most sensitive readouts available. Our results indicate that the antigen sensitivity of model systems used in research such as Jurkat cells, or in the clinic (primary T cells) are up to several orders of magnitude lower than that of the parental T-cell clone, and that the sensitivity of the latter can be accurately replicated by combining TCR transfer with endogenous TCR knockout (TCR replacement) in primary T cells. Apart from having implications for designing more effective TCR-based immunotherapies, this result indicates that TCR replacement is preferable to TCR transfer for functional characterizations of TCRs of interest especially where these TCRs compete poorly with endogenous TCRs for surface expression or have a relatively low affinity for cognate antigen. This approach should also enable detailed analysis of TCR recognition in the absence of parental T-cell clones. Such TCRs may come from T cells that display poor growth characteristics (eg, as a result of cancer-mediated T-cell exhaustion57 ), or directly from high-throughput sequencing of TCR repertoires.58 One can also envisage that primary T cells transduced with a TCR of unknown specificity but not expressing the endogenous TCRs could be used for high-throughput, whole-genome screens28 to identify new TCR ligands, and therefore new potential therapeutic targets.
γδ T cells offer an attractive tool for cancer immunotherapy, due to their ability to recognize ubiquitously expressed targets and no evidence of MHC restriction. This feature allows such γδ T cells to respond to cancer from any individual and also eliminates the risk of graft-versus-host disease.59 To date, the majority of clinical trials utilizing γδ T cells have focused on the predominant subset in the periphery, namely Vγ9Vδ2 T cells, which respond to phosphoantigen metabolites. Several multicenter clinical trials60-63 have demonstrated that in vivo activation of Vγ9Vδ2 T cells and cancer cell sensitization with aminobisphosphonates (zoledronate, pamidronate) was well tolerated and did not result in off-target toxicities (despite the ubiquitous expression of butyrophilin molecules and mevalonate pathway components). Encouragingly, aminobisphosphonate treatment resulted in objective clinical responses in a fraction of patients with non-Hodgkin lymphoma, multiple myeloma,60 and AML,62 demonstrating the potential of γδ T-cell–based immunotherapies for hematological malignancies. However, the therapeutic success of γδ T-cell immunotherapies remains underwhelming,11 especially compared with CD19-CAR therapies.64 One of the potential reasons for this poor success could be the use of a variable and largely undefined (especially in terms of TCR usage) cellular product. Furthermore, antigen-driven expansion of Vγ9Vδ2 T cells, as used so far, has been shown to lead to exhaustion and loss of functional activity in both animal models and in patients.65,66 In contrast, TCR replacement by gene transfer, as used here, could be applied to a desirable T-cell subset (for instance, T memory stem cells) thereby potentially allowing improved host engraftment and/or function.67 We propose that using a defined γδ TCR transferred to patient’s T cells in combination with the knockout of endogenous αβ TCRs could be a therapeutically beneficial strategy. Indeed, TCR-plus-CRISPR T cells showed a markedly stronger response (in terms of TNFα and IFNγ production) than TCR-only transduced T cells to established cancer cell lines, as well as all primary B-ALL blasts. It should be noted that TNFα production was shown to correlate with cancer-specific activity of cytotoxic T cells, and an elevated intratumoral TNFα concentration could serve as a favorable prognostic factor.68 Similarly, IFNγ is a potent immunomodulatory cytokine that enhances T-cell–mediated recognition of cancer cells and plays a direct anticancer role.69,70 No on-target off-tumor reactivity of γδ20 TCR-transduced cells was observed against primary, aminobisphosphonate-treated B cells, T cells, or whole PBMCs, in line with multiple clinical trials indicating that aminobisphosphonate treatment (with or without infusion of ex vivo–expanded γδ T cells) was generally well tolerated without severe adverse effects (reviewed in Legut et al11 ). It should be noted, however, that even with using the TCR replacement technology for generation of Vγ9Vδ2 TCR-transgenic T cells, the patients will most likely require bisphosphonate treatment for efficient cancer sensitization, in addition to adoptive transfer of TCR-engineered T cells. The increasing clinical experience in coadministering aminobisphosphonates and Vγ9Vδ2 T cells62,71-74 will undoubtedly facilitate designing of clinical trials testing the efficacy of Vγ9Vδ2 TCR-transgenic T cells. We therefore believe that the TCR replacement technology described here would be of use in fundamental and translational research where it could, for instance, be used to discover ligands of clinically relevant TCRs. In addition, this kind of approach has potential for developing a new generation of TCR-based immunotherapies, provided the method is optimized for the clinical scale, based on the wealth of experience in generating therapeutic CAR T cells. Widespread clinical application of gene-editing technology seems imminent, as demonstrated by the recent success of the off-the-shelf allogeneic CAR19 T cells in inducing remission of B-ALL in infants.53 In summary, it seems likely that TCR replacement by CRISPR/Cas9, or other means, will generate clinically useful T cells that do not encompass the dangers of TCR mispairing and that can be orders of magnitude more sensitive than the products currently being trialed.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Catherine Naseriyan from Central Biotechnology Services for the assistance with cell sorting.
M.L. was supported by the Cancer Research UK scholarship. A.K.S. is a Wellcome Trust Senior Investigator (WT100327MA).
Authorship
Contribution: M.L., G.D., and A.K.S. conceived the study, designed the experiments, and wrote the manuscript; M.L. performed the experiments and analyzed the data; and A.A.M. and O.G.O. provided the reagents and guidance.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew K. Sewell, Cardiff University, Henry Wellcome Building, Heath Park, CF14 4XN Cardiff, United Kingdom; e-mail: sewellak@cardiff.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal