Key Points
FOXO1 activity is essential for growth and maintenance of BCP-ALL.
Inhibition of FOXO1 reduces leukemia load and prolongs survival in a preclinical model of BCP-ALL.
Abstract
The FOXO1 transcription factor plays an essential role in the regulation of proliferation and survival programs at early stages of B-cell differentiation. Here, we show that tightly regulated FOXO1 activity is essential for maintenance of B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Genetic and pharmacological inactivation of FOXO1 in BCP-ALL cell lines produced a strong antileukemic effect associated with CCND3 downregulation. Moreover, we demonstrated that CCND3 expression is critical for BCP-ALL survival and that overexpression of CCND3 protected BCP-ALL cell lines from growth arrest and apoptosis induced by FOXO1 inactivation. Most importantly, pharmacological inhibition of FOXO1 showed antileukemia activity on several primary, patient-derived, pediatric ALL xenografts with effective leukemia reduction in the hematopoietic, lymphoid, and central nervous system organ compartments, ultimately leading to prolonged survival without leukemia reoccurrence in a preclinical in vivo model of BCP-ALL. These results suggest that repression of FOXO1 might be a feasible approach for the treatment of BCP-ALL.
Introduction
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) represents the most common malignancy in children originating from pro– and pre–B cells.1 Although curable in most cases, treatment of high-risk and relapsed BCP-ALL remains a therapeutic challenge.2 Therefore, novel strategies targeting the oncogenic program of leukemia cells are of particular interest.
In BCP-ALL, alterations in survival, proliferation, and differentiation programs in the cells of origin have been described.3 For instance, in pro–B ALL (no pre–B-cell receptor expression, pre-BCR−) activating mutations and translocations of cytokine signaling pathways contribute to leukemia. Similarly, in pre-BCR+ BCP-ALL the oncogenic activation of pre-BCR-dependent proliferation and survival pathways including BCR-ABL1 fusion have been described.3 In both types of BCP-ALL, constitutive activation of key prosurvival pathways including phosphatidylinositol 3-kinase (PI3K)–AKT, MAPK/extracellular signal-regulated kinase (ERK), NF-κB, and JAK/STAT is essential and can be targeted by pharmacological inhibitors.4 Three of these pathways (ie, PI3K-AKT, MAPK/ERK, and NF-κB) are known to inactivate FOXO1, a master regulator of pro– and pre–B-cell differentiation.5-8
FOXO1 is a member of the forkhead family of transcription factors, also including FOXO3, FOXO4, and FOXO6, which all share a highly conserved forkhead DNA binding domain but differ in tissue specificity.9 FOXO transcription factors are partially redundant regulators of growth arrest, cell death, protection against oxidative stress, and metabolism.10,11 Posttranslational modifications define subcellular localization and protein-protein interactions of FOXOs, ultimately regulating their transcriptional activity.10-12 In BCP-ALL, FOXO1 activation induces growth arrest and apoptosis.6 Indeed, FOXO1 is phosphorylated at AKT-specific sites in pre-BCR+6 and in BCR-ABL1+ BCP-ALL cell lines.7 Pharmacological inactivation of pre-BCR signaling by SYK- or ABL1-inhibitors or overexpression of constitutive active FOXO1 leads to apoptosis.6
Paradoxically, despite phosphorylation, a substantial proportion of FOXO1 is localized in nuclei6,7 and apparently activates transcription of direct FOXO targets including RAG1, RAG2, AICDA, and the pre-BCR component VPREB1, which are all essential for leukemogenesis and clonal evolution of BCP-ALL,7,13,14 contradicting the alleged tumor suppressor role of FOXO1.
Importantly, FOXO1 is an essential component of the proliferation and survival program at early stages of B-cell differentiation.15,16 Moreover, FOXO-inactivating pathways are tightly regulated in pre–B-cell ALL. Specifically, knockdown of PTEN, a PI3K-AKT inhibitor, or expression of constitutive active versions of AKT or SYK prevented leukemia development in a BCR-ABL1+ BCP-ALL model repressing leukemia growth.17 Similarly, depletion of negative feedback regulators of ERK (Spry2, Dusp6, and Etv5) repressed pre–B-cell leukemogenesis.18
Therefore, we hypothesized that a tight regulation of FOXO1 might be critical for maintenance of BCP-ALL.19 Using genetic and pharmacological approaches inactivating FOXO1 in BCP-ALL, we investigated the antileukemia activity and molecular consequences of FOXO1 depletion. Using ex vivo and in vivo models of patient-derived BCP-ALL, we demonstrated the efficacy of pharmacological FOXO1 inactivation pointing toward a novel strategy to treat BCP-ALL.
Methods
Additional and detailed information on methods is provided in the supplemental Methods (available on the Blood Web site).
NOD/SCID/huALL
In this study, we used 10 patient-derived xenograft (PDX) samples established by transplantation of patient ALL cells onto NOD/SCID mice (NOD.CB17-Prkdcscid/J, Charles River) as previously described.20,21 Patient samples were obtained at initial diagnosis or second relapse after informed consent in accordance with the institution’s (Ulm University) ethical review board. All animal experiments were approved by the appropriate authority (Regierungspräsidium Tübingen). For ex vivo experiments, PDX leukemia samples were isolated from spleens of ALL-bearing mice and exposed to AS1842856. Cell death was analyzed (according to forward-scattered/side-scattered light [FSC/SSC] criteria) by flow cytometry. For in vivo analyses, recipients were transplanted with primograft samples, and upon leukemia manifestation (≥5% huCD19+ cells in peripheral blood as detected by flow cytometry),22,23 randomly grouped receiving either AS1842856 or dimethyl sulfoxide (DMSO). After treatment, animals were either euthanized estimating leukemia loads in spleen, bone marrow, or central nervous system or further followed up until reoccurrence of leukemia. PDX characteristics are summarized in supplemental Table 1B.
Cell lines
BCP-ALL cell lines KOPN-8, REH, NALM-6, RS4;11, BV-173, RCH-ACV, and EU-3; T-cell acute lymphoblastic leukemia (T-ALL) lines Jurkat, CCRF-CEM, MOLT-4, BE-13, and ALL-SIL; diffuse large B-cell lymphoma (DLBCL) lines HBL-1, OCI-LY3, and OCI-LY5; follicular lymphoma (FL) lines DOHH2, WSU-FSCCL, WSU-NHL, and Karpas-422; and classical Hodgkin lymphoma (cHL) lines SUP-HD1 and L428 were purchased from DSMZ (Braunschweig, Germany). The BCP-ALL cell line UoCB6 was kindly provided by J. D. Rowley (Chicago, IL). The 018Z BCP-ALL cell line was described earlier.24 The U-HO1 cell line was obtained from the Institute of Pathology, Ulm University, Ulm, Germany.25 All cell lines were cultured as described earlier.20,26 Cell line identity and mycoplasma contamination were controlled regularly.
Vectors and lentiviral transduction
Lentiviral supernatants were produced and cell lines were transduced as described.27 FOXO1 and CCND3 targeting short hairpin RNAs (shRNAs) and scrambled control shRNA were expressed using the pRSI12-U6-sh-UbiC-TagRFP-2A-Puro lentiviral vector (BioCat, Heidelberg, Germany). For expression of genes, we used the SF-LV-cDNA-eGFP vector.28
Flow cytometry and cell sorting
Growth dynamics of cell lines transduced with a lentiviral vector coexpressing red fluorescent protein (RFP) were monitored by flow cytometry (FACSCalibur, BD Biosciences). For biochemical analysis, RFP+ cells were sorted by the Flow Cytometry Core Facility using FACSAria cell sorter (BD Biosciences). The cell cycle distribution was measured by flow cytometry using propidium iodide (PI), and cell death was assessed by annexin-V/PI/7-aminoactinomycin staining as described.26
Cell viability analysis
Cell viability was analyzed by cell counting using a Vi-cell XR cell viability analyzer based on trypan blue exclusion (Beckman Coulter, Krefeld, Germany). Fifty percent inhibitory concentration (IC50) values were calculated by fitting the data points to a nonlinear regression curve using GraphPad Prism (GraphPad Software, San Diego, CA).
Gene expression analysis
Total RNA was extracted and gene expression profiles were analyzed using Human Gene 1.0 ST Affymetrix GeneChip arrays as described previously.27 Probe-level data were obtained using the robust multichip average normalization algorithm. The analysis of differentially expressed genes was achieved (GeneSifter microarray data analysis software, http://www.genesifter.net; PerkinElmer, Waltham, MA). For all cell lines and conditions, 2 biological replicates were analyzed. To identify pathways significantly modulated by FOXO1 depletion, we used gene set enrichment analysis (GSEA; http://software.broadinstitute.org/gsea/index.jsp). Microarray data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/gds/; accession number GSE86067).
Statistical analysis
GraphPad Prism 5 was used for all statistical analyses. The data are expressed as mean ± standard deviation (SD). A 2-tailed Student t test was used to compare the differences between 2 groups. Differences were also evaluated by using the standard 1-way analysis of variance (ANOVA) followed by Bonferroni post hoc test in case of >2 groups. P < .05 was considered to be statistically significant. For analysis of animal experiments, we used the Mann-Whitney U test. For Kaplan-Meier analysis, we used log-rank test.
Results
BCP-ALL cells constitutively express nuclear FOXO1 despite activated AKT signaling
In pre-BCR+ or BCR/ABL+ BCP-ALL, an AKT-dependent inactivating phosphorylation of FOXO1 protein was reported.6,7 We therefore addressed FOXO1 expression and activity in 4 pre-BCR+ (NALM6, O18Z, KOPN-8, and EU3/697) and 4 pre-BCR− cell lines (REH, RS4;11, MUTZ-5, and UoCB6). FOXO1 was expressed in all lines, although the expression levels differed (Figure 1A). We also analyzed FOXO1 expression in PDXs (pre-BCR−, PDX#1, 2, 3, 4, 5, 6, 9; pre-BCR+, #10; supplemental Table 1B) and found FOXO1 expression levels comparable to the BCP-ALL cell lines independently of pre-BCR expression (Figure 1B). Because FOXO3A messenger RNA (mRNA) expression ranks second after FOXO1 in pro– and pre–B cells,6 we also analyzed FOXO3A protein levels in BCP-ALL cell lines (Figure 1A) and compared levels of FOXO1 and FOXO3A mRNA expression in BCP-ALL cell lines and PDXs (Figure 1C). The qRT-PCR analyses demonstrated relatively higher FOXO1 mRNA levels in most BCP-ALL cell lines and PDXs. AKT-dependent phosphorylation of FOXO1 (positions T24 and S256) was detected in all cell lines, independently of pre-BCR expression and with intensities similar to total FOXO1 (Figure 1A). We therefore assessed localization of FOXO1 by subcellular fractionation (Figure 1D) and by immunofluorescence (Figures 1E). Interestingly, both methods confirmed presence of FOXO1 in the nuclei independent of the pre-BCR status.
Expression of FOXO1 in BCP-ALL. (A) Expression of FOXO1 and FOXO3A proteins and phosphorylation of FOXO1 in BCP-ALL cell lines analyzed by immunoblot. (B) Expression of FOXO1 in PDX ALL analyzed by immunoblot. (C) Relative expression of FOXO1 and FOXO3A mRNA in BCP-ALL cell lines and PDX ALL samples. FOXO1 and FOXO3 mRNA expression was analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The log2 of FOXO1/FOXO3A ratio was calculated as CtFOXO3a-CtFOXO1, where Ct is cycle threshold. Data are shown as mean ± SD (N = 3). (D) Subcellular distribution of FOXO1. After subcellular fractionation, expression of FOXO1 was detected by immunoblot; purity of cytoplasmatic and nuclear fractions was controlled by expression of tubulin B (TUBB) and histone 3 (H3), respectively. (E) Immunofluorescence analysis of FOXO1 localization in BCP-ALL cell lines. Cells were attached to the surface of glass slides by low-speed centrifugation and stained by anti-FOXO1 antibody (green). Nuclei were counterstained with PI (red). Images were acquired using a Zeiss MTB2004 system using Axiovision software (63× oil immersion lens, original magnification ×228). (F) Expression of inducible constitutive-active FOXO1 (FOXO1(3A)ER in BCP-ALL cell lines (F1ER) and wild-type FOXO1 (wtFOXO1). (G) FOXO1 activation is toxic for BCP-ALL cell lines. Pre-BCR+ (018Z and RCH-ACV) and Pre-BCR− (UoCB6) cell lines transduced with FOXO1(3A)ER or with control vector were sorted and treated with 200 nM 4-OHT. After 48 hours, live cells were counted using trypan blue exclusion. The data are presented as percentage of control nontreated cells (mean ± SD). The significance by 2-sided Student t test (N = 3). ****P < .0001.
Expression of FOXO1 in BCP-ALL. (A) Expression of FOXO1 and FOXO3A proteins and phosphorylation of FOXO1 in BCP-ALL cell lines analyzed by immunoblot. (B) Expression of FOXO1 in PDX ALL analyzed by immunoblot. (C) Relative expression of FOXO1 and FOXO3A mRNA in BCP-ALL cell lines and PDX ALL samples. FOXO1 and FOXO3 mRNA expression was analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The log2 of FOXO1/FOXO3A ratio was calculated as CtFOXO3a-CtFOXO1, where Ct is cycle threshold. Data are shown as mean ± SD (N = 3). (D) Subcellular distribution of FOXO1. After subcellular fractionation, expression of FOXO1 was detected by immunoblot; purity of cytoplasmatic and nuclear fractions was controlled by expression of tubulin B (TUBB) and histone 3 (H3), respectively. (E) Immunofluorescence analysis of FOXO1 localization in BCP-ALL cell lines. Cells were attached to the surface of glass slides by low-speed centrifugation and stained by anti-FOXO1 antibody (green). Nuclei were counterstained with PI (red). Images were acquired using a Zeiss MTB2004 system using Axiovision software (63× oil immersion lens, original magnification ×228). (F) Expression of inducible constitutive-active FOXO1 (FOXO1(3A)ER in BCP-ALL cell lines (F1ER) and wild-type FOXO1 (wtFOXO1). (G) FOXO1 activation is toxic for BCP-ALL cell lines. Pre-BCR+ (018Z and RCH-ACV) and Pre-BCR− (UoCB6) cell lines transduced with FOXO1(3A)ER or with control vector were sorted and treated with 200 nM 4-OHT. After 48 hours, live cells were counted using trypan blue exclusion. The data are presented as percentage of control nontreated cells (mean ± SD). The significance by 2-sided Student t test (N = 3). ****P < .0001.
Attenuated FOXO1 activity is critical for BCP-ALL survival
Analysis of published data on sensitivity of tumor cell lines to pharmacological inhibition of AKT revealed that BCP-ALL cell lines are sensitive to the pan-AKT inhibitor GSK690693.29 Importantly, there were no differences between pre-BCR+ and pre-BCR− cells in terms of sensitivity to GSK690693 (supplemental Table 2), MK-2206, and the PI3Kα/δ inhibitor pictilisib (supplemental Table 3; Genomics of Drug Sensitivity in Cancer database, http://www.cancerrxgene.org/).30
Previously, sensitivity of pre-BCR+ cell line RCH-ACV toward expression of an AKT-resistant inducible version of FOXO1 (FOXO1(3A)ER) has been shown.6 We extended this study with additional pre-BCR+ (018Z) and pre-BCR− (UoCB6) cell lines. In all cell lines, expression of FOXO1(3A)ER significantly decreased cell viability compared with control groups (Figure 1F-G).
Thus, constitutive activation of AKT attenuating FOXO1 levels is essential for BCP-ALL maintenance independently of pre-BCR expression. Nevertheless, despite AKT activation a substantial proportion of FOXO1 is located in the nuclei indicating a role of FOXO1 in BCP-ALL.
FOXO1 expression is essential for BCP-ALL cell lines
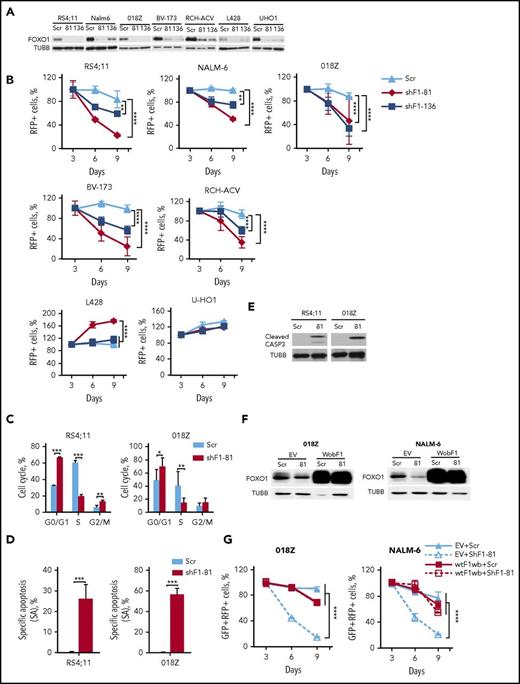
We investigated the importance of FOXO1 using shRNA-expressing lentiviral vectors that harbor the fluorescent marker RFP. We selected 2 shRNA constructs that efficiently depleted FOXO1: shFOXO1-81 specifically targeting FOXO1 only and shFOXO-136, which in addition to FOXO1 also targets FOXO3A and FOXO631 (Figure 2A). We assessed the effect of FOXO1 repression by monitoring the proportions of RFP+ cells over time. FOXO1 shRNAs decreased the RFP+ population in BCP-ALL cell lines in a time-dependent manner (Figure 2B).
FOXO1 knockdown induces growth arrest and apoptosis in BCP-ALL. (A) Efficient shRNA-mediated knockdown of FOXO1. BCP-ALL and cHL (L428, U-HO1) cell lines were transduced with lentiviral vectors expressing scrambled (Scr), FOXO1-targeting shRNA-81 (shF1-81) or shRNA targeting FOXO1, FOXO3A, and FOXO6 (shF1-136). Transduced cells were selected by incubation with puromycin, and FOXO1 expression was assessed by immunoblot. A representative of 2 independent experiments is shown. (B) Antileukemia effect of FOXO1 knockdown. The dynamic of the RFP+ population was measured every 3 days starting at day 3 after transduction. The proportion of RFP+ cells on first measurement (day 3) was taken as 100%. Data represent mean ± SD of 3 independent transductions. (C-E) Cells transduced with Scr or shRF1-81 vectors were sorted on day 4 after transduction. On day 5 cell cycle phase distribution (C) and cell death (D) were measured by PI and by annexin V–fluorescein isothiocyanate/PI staining, respectively. Mean percentages ± SD, 2 independent experiments, each measured twice (C). Specific apoptosis was calculated as SA(%) = 100 × (AE − AC)/(100 − AC), where AE equals the percentage of apoptotic cells in the experimental group and AC equals the percentage of apoptotic cells in the control group (N = 3) (D). (E) Induction of caspase-3 cleavage by FOXO1 depletion. Cleaved CASP3 was detected by immunoblot in lysates of sorted BCP-ALL cell lines. A representative image of 2 independent experiments is shown. (F-G) Overexpression of wild-type FOXO1 harboring wobbled shRNA-targeted sequence (wtF1wb) protects 018Z and NALM-6 cells from the cytotoxic effect of shF1-81. Cells were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-wtF1wb-expressing vector. In 4 to 6 days, the cells were transduced with shF1-81 or Scr vectors expressing the fluorescent marker RFP. For FOXO1 protein analysis, cells were sorted by fluorescence-activated cell sorting (FACS) on day 3 after the second transduction (F). The percentage of the GFP+/RFP+ population was measured at indicated time points after second transduction. Data are shown as mean ± SD of 2 independent experiments (G). *P < .05; **P < .01; ***P < .001; ****P < .0001.
FOXO1 knockdown induces growth arrest and apoptosis in BCP-ALL. (A) Efficient shRNA-mediated knockdown of FOXO1. BCP-ALL and cHL (L428, U-HO1) cell lines were transduced with lentiviral vectors expressing scrambled (Scr), FOXO1-targeting shRNA-81 (shF1-81) or shRNA targeting FOXO1, FOXO3A, and FOXO6 (shF1-136). Transduced cells were selected by incubation with puromycin, and FOXO1 expression was assessed by immunoblot. A representative of 2 independent experiments is shown. (B) Antileukemia effect of FOXO1 knockdown. The dynamic of the RFP+ population was measured every 3 days starting at day 3 after transduction. The proportion of RFP+ cells on first measurement (day 3) was taken as 100%. Data represent mean ± SD of 3 independent transductions. (C-E) Cells transduced with Scr or shRF1-81 vectors were sorted on day 4 after transduction. On day 5 cell cycle phase distribution (C) and cell death (D) were measured by PI and by annexin V–fluorescein isothiocyanate/PI staining, respectively. Mean percentages ± SD, 2 independent experiments, each measured twice (C). Specific apoptosis was calculated as SA(%) = 100 × (AE − AC)/(100 − AC), where AE equals the percentage of apoptotic cells in the experimental group and AC equals the percentage of apoptotic cells in the control group (N = 3) (D). (E) Induction of caspase-3 cleavage by FOXO1 depletion. Cleaved CASP3 was detected by immunoblot in lysates of sorted BCP-ALL cell lines. A representative image of 2 independent experiments is shown. (F-G) Overexpression of wild-type FOXO1 harboring wobbled shRNA-targeted sequence (wtF1wb) protects 018Z and NALM-6 cells from the cytotoxic effect of shF1-81. Cells were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-wtF1wb-expressing vector. In 4 to 6 days, the cells were transduced with shF1-81 or Scr vectors expressing the fluorescent marker RFP. For FOXO1 protein analysis, cells were sorted by fluorescence-activated cell sorting (FACS) on day 3 after the second transduction (F). The percentage of the GFP+/RFP+ population was measured at indicated time points after second transduction. Data are shown as mean ± SD of 2 independent experiments (G). *P < .05; **P < .01; ***P < .001; ****P < .0001.
Because we have shown that FOXO1 acts as a tumor suppressor in cHL,26 we used the cHL cell lines L428 and U-HO1 as a negative control. Both FOXO1 shRNA constructs reduced FOXO1 expression (Figure 2A) but did not reduce the proportion of RFP+ cells (U-HO1) or even stimulated proliferation (L428) in cHL cell lines (Figure 2B). We addressed the effect of FOXO1 knockdown on cell-cycle progression and apoptosis in RS4;11 (pre-BCR−) and 018Z (pre-BCR+) cell lines. Knockdown of FOXO1 resulted in G1-arrest and induction of apoptosis in both lines (Figure 2C-D), which was associated with caspase-3 cleavage (Figure 2E).
To exclude off-target effects of RNA interference, we performed a rescue experiment and overexpressed wild-type FOXO1 (wtFOXO1) harboring wobbled nucleotides (wtFOXO1wb) at the shFOXO1-81-target site in 018Z and NALM-6 cells (Figure 2F). In contrast to the AKT-insensitive inducible FOXO1 construct, expression of wtFOXO1wb only slightly influenced proliferation of the cells. However, wtFOXO1wb protected the cells from FOXO1-targeting shRNA (Figure 2G).
Thus, BCP-ALL cells depend on expression of FOXO1.
FOXO1 knockdown represses CCND3 and mechanistic target of rapamycin (mTOR) activity in BCP-ALL
Given that knockdown of FOXO1 induced both cell cycle arrest and apoptosis, we measured expression of the FOXO1 target gene cyclin D3 (CCND3), which is essential for proliferation of pro– and pre–B cells32 and mediates antiapoptotic functions.33 FOXO1 knockdown led to repression of CCND3 at mRNA and protein levels (Figure 3A-B). Knockdown of CCND3 by itself had a strong cytotoxic effect in both RS4;11 and 018Z cell lines (Figure 3C-D), which was associated with decreased expression of the proliferation-related protein MKI67, G1-cell cycle arrest, and caspase-3 cleavage (Figure 3E-G). CCND3 knockdown repressed RB1 phosphorylation, attenuated MYC expression, and increased expression of CDKN1B/p27 (Figure 3G).
FOXO1 knockdown downregulates CCND3 transcription. (A-B) RS4;11 and 018Z BCP-ALL cell lines were transduced with lentiviral vectors expressing scrambled (Scr) or FOXO1-specific shRNA-81. Cells were sorted by FACS on day 5 after transduction and used for isolation of total RNA and protein. Downregulation of CCND3 mRNA (A) and protein (B) by shFOXO1-81 were measured by qRT-PCR (N = 2) and immunoblot, respectively. A representative of 2 independent protein isolations is shown. (C) Efficient knockdown of CCND3. BCP-ALL cell lines were transduced with a lentiviral vector expressing scrambled (Scr) or targeting CCND3 (shC3) shRNA. The RFP+ cells were sorted by FACS on day 3 after transduction and CCND3 expression was assessed by immunoblot. A representative of 2 independent experiments is shown. (D) Antileukemia effect of CCND3 knockdown. The dynamic of the RFP+ population was measured every 3 days starting from the day 3 after infection. The proportion of RFP+ cells on the first day of measurement (day 3) was taken as 100%. Data represent mean ± SD of 3 independent transductions. (E) CCND3 knockdown decreases expression of the proliferation marker MKI67. Cells were sorted on day 3 after transduction and used for protein isolation. Repression of MKI67 was demonstrated by immunoblot. (F) CCND3 knockdown induces G1 cell cycle arrest. Transduced cells were sorted on day 3 after transduction, and cell cycle was analyzed by PI staining on the next day after sorting (N = 2). (G) CCND3 knockdown downregulates RB1 phosphorylation and MYC expression, induces CASP3 cleavage, and increases CDKN1B expression. Cells were sorted on day 3 after transduction and the protein expression and phosphorylation was measured by immunoblot (N = 2). (H-I) CCND3 overexpression protects BCP-ALL cells from the cytotoxic effect of FOXO1 knockdown. 018Z and NALM-6 cells were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-CCND3-expressing vector. In 4 to 6 days, the cells were transduced with shF1-81 or scr vectors expressing the fluorescent marker RFP. (H) For analysis of CCND3 protein expression, the cells were sorted by FACS on day 3 after the second transduction. (I) The percentage of the GFP+/RFP+ population was measured at indicated time points after the second transduction. Data are shown as mean ± SD (N = 2). (J) CCND3 overexpression restores RB1 phosphorylation and MYC expression, but not the decrease of S6 phosphorylation induced by FOXO1 knockdown (N = 2). The 018Z cells overexpressing CCND3 of control vector were transduced with vectors expressing shF1-81 or scr and sorted on day 4 after transduction. The protein expression and phosphorylation status was measured by immunoblot (N = 2). *P < .05; **P < .01; ***P < .001; ****P < .0001.
FOXO1 knockdown downregulates CCND3 transcription. (A-B) RS4;11 and 018Z BCP-ALL cell lines were transduced with lentiviral vectors expressing scrambled (Scr) or FOXO1-specific shRNA-81. Cells were sorted by FACS on day 5 after transduction and used for isolation of total RNA and protein. Downregulation of CCND3 mRNA (A) and protein (B) by shFOXO1-81 were measured by qRT-PCR (N = 2) and immunoblot, respectively. A representative of 2 independent protein isolations is shown. (C) Efficient knockdown of CCND3. BCP-ALL cell lines were transduced with a lentiviral vector expressing scrambled (Scr) or targeting CCND3 (shC3) shRNA. The RFP+ cells were sorted by FACS on day 3 after transduction and CCND3 expression was assessed by immunoblot. A representative of 2 independent experiments is shown. (D) Antileukemia effect of CCND3 knockdown. The dynamic of the RFP+ population was measured every 3 days starting from the day 3 after infection. The proportion of RFP+ cells on the first day of measurement (day 3) was taken as 100%. Data represent mean ± SD of 3 independent transductions. (E) CCND3 knockdown decreases expression of the proliferation marker MKI67. Cells were sorted on day 3 after transduction and used for protein isolation. Repression of MKI67 was demonstrated by immunoblot. (F) CCND3 knockdown induces G1 cell cycle arrest. Transduced cells were sorted on day 3 after transduction, and cell cycle was analyzed by PI staining on the next day after sorting (N = 2). (G) CCND3 knockdown downregulates RB1 phosphorylation and MYC expression, induces CASP3 cleavage, and increases CDKN1B expression. Cells were sorted on day 3 after transduction and the protein expression and phosphorylation was measured by immunoblot (N = 2). (H-I) CCND3 overexpression protects BCP-ALL cells from the cytotoxic effect of FOXO1 knockdown. 018Z and NALM-6 cells were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-CCND3-expressing vector. In 4 to 6 days, the cells were transduced with shF1-81 or scr vectors expressing the fluorescent marker RFP. (H) For analysis of CCND3 protein expression, the cells were sorted by FACS on day 3 after the second transduction. (I) The percentage of the GFP+/RFP+ population was measured at indicated time points after the second transduction. Data are shown as mean ± SD (N = 2). (J) CCND3 overexpression restores RB1 phosphorylation and MYC expression, but not the decrease of S6 phosphorylation induced by FOXO1 knockdown (N = 2). The 018Z cells overexpressing CCND3 of control vector were transduced with vectors expressing shF1-81 or scr and sorted on day 4 after transduction. The protein expression and phosphorylation status was measured by immunoblot (N = 2). *P < .05; **P < .01; ***P < .001; ****P < .0001.
In order to determine a role of CCND3 in the antileukemic effect of FOXO1 knockdown, we cotransduced 018Z and NALM-6 cells with shFOXO1-81(RFP+) and CCND3(GFP+)-expressing vectors, or with control empty vector Scr(RFP+) and (EV)(GFP+) vectors. CCND3 overexpression did not influence proliferation of control cells, but completely blocked the cytotoxic effect of FOXO1 knockdown (Figure 3H-I).
Finally, we treated T- and BCP-ALL lines with palbociclib, an inhibitor of the cyclin d-dependent kinases CDK4/6. Because CCND3 is a core survival factor of T-ALL and the T-ALL cell lines are highly sensitive to palbociclib,34 we included them as control. All cell lines responded similarly with a G1-arrest and increased apoptosis (supplemental Figure 1). Further, we compared sensitivities of additional BCP- and T-ALL lines to palbociclib by mining the Genomics of Drug Sensitivity in Cancer database. Although the IC50 values of T- and B-ALLs did not differ significantly, proportionally more BCP-ALLs showed sensitivity (IC50 <1 µM) to the inhibitor. However, pre-BCR expression was not associated with palbociclib sensitivity (supplemental Table 4). Interestingly, B-ALL and T-ALL lines are equally sensitive to the other CDK4/6 inhibitor LEE011/Ribociclib.35
FOXO1 knockdown inhibits mTOR activity and MYC expression in human umbilical vein endothelial cells (HUVECs).36 Of note, RICTOR, a subunit of mTORC2, is a conserved FOXO target.37 Respectively, FOXO1 knockdown by shFOXO1-81 downregulated MYC and RICTOR mRNA and protein expression (supplemental Figure 2A-D). Downregulation of RICTOR (supplemental Figure 2C) was associated with decrease of mTORC2 activity indicated by decreased AKT S473 phosphorylation (supplemental Figure 2D). Additionally, FOXO1 knockdown decreased expression of mTOR kinase and phosphorylation of ribosomal S6 kinase (S6K), S6 protein, and inhibitor of translation EIF4EBP1 (supplemental Figure 2D). Thus, knockdown of FOXO1 in BCP-ALL consistently downregulated mTOR activity. FOXO1 knockdown by shFOXO1-136, which inhibited FOXO1 to a somewhat lesser extent, also downregulated expression of RICTOR, MYC, and CCND3 (supplemental Figure 2E).
We asked whether rescue of BCP-ALL cells from FOXO1 knockdown by CCND3 overexpression is associated with restoration of MYC expression and mTORC1 activation. RB1 phosphorylation was used as control of FOXO1 depletion and CCND3 activity. We found that CCND3 overexpression restores expression of MYC, but not mTORC1 activity (Figure 3J). Finally, we investigated effects of RICTOR and MYC knockdown on CCND3 expression. For RICTOR knockdown we selected 2 most effective shRNA constructs. For MYC knockdown we used a well-described specific shRNA.38 Knockdown of RICTOR did not influence expression of CCND3 (supplemental Figure 3A), whereas knockdown of MYC slightly decreased CCND3 levels (supplemental Figure 3B).
Thus, downregulation of CCND3 contributes to the cytotoxic effect of FOXO1 depletion in BCP-ALL.
BCP-ALL cell lines are sensitive to pharmacological FOXO1 inhibition
Next, we investigated the feasibility of pharmacological FOXO1 inhibition to treat BCP-ALL and analyzed sensitivities of BCP-, T-ALL, B-cell NHL, and cHL cell lines to the small molecular weight FOXO1 inhibitor AS1842856. The BCP-ALL cells showed highest sensitivity to AS1842856 (Figure 4A), which did not depend on pre-BCR expression (Figure 4B). Moreover, we investigated kinetics and mechanisms of AS1842856 in the 3 most sensitive cell lines. Because AS1842856 plasma concentrations after oral administration in mice reaches 300 nM,39 we used the inhibitor at a range of 20 to 320 nM and observed a dose-dependent decrease of live cells and increasing apoptosis induction (Figure 4C-E). Cell cycle analysis revealed G0/G1-arrest in BCP-ALL but not in cHL control cell lines (supplemental Figure 4).
BCP-ALL cell lines are explicitly sensitive to the FOXO1 inhibitor AS1842856. (A) Comparative analysis of sensitivities of leukemia and lymphoma cell lines to AS1842856. Cells were incubated for 6 days with increasing concentrations of AS1842856. The antitumor effect was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, and IC50 values were calculated. (B) Comparison of the sensitivities of pre-BCR− and pre-BCR+ BCP-ALL cells to AS1842856 (P = .400, 2-sided Student t test). AS1842856 decreases viability (C) and induces apoptosis (D) in BCP-ALL cells in a time- and dose-dependent manner. Cells were incubated in the presence of increasing concentrations of AS1842856. The cell number and viability was measured by trypan blue exclusion at indicated time points. Apoptosis was measured using annexin V/PI staining. Data are shown as means of specific cell death ± SD (N = 3). (E) CASP3 cleavage, detection by immunoblot (RS4;11 and 018Z, 40 nM; UoCB6, 80 nM AS1842856), a representative of 3 independent experiments is shown. (F-G) NALM-6 cells were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-wtFOXO1 (WTF1)–expressing vector. For protein analysis, cells were sorted 3 days after transduction (F). (G) Four to 6 days after transduction cells were treated with AS1842856. Percentages of GFP+ cells (by flow cytometry) and the total number of live cells (by cell counting) were measured at indicated time points. Data are shown as fold changes normalized to the initial number of live GFP+ cells. The number of live GFP+ cells was calculated as N × %GFP+ cells/100, where N is the number of live cells per well and %GFP+ is the percentage of GFP+ cells. Data are shown as mean ± SD of 3 independent experiments. ANOVA, *P < .05; **P < .01; ***P < .001, and ****P < .0001, respectively.
BCP-ALL cell lines are explicitly sensitive to the FOXO1 inhibitor AS1842856. (A) Comparative analysis of sensitivities of leukemia and lymphoma cell lines to AS1842856. Cells were incubated for 6 days with increasing concentrations of AS1842856. The antitumor effect was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, and IC50 values were calculated. (B) Comparison of the sensitivities of pre-BCR− and pre-BCR+ BCP-ALL cells to AS1842856 (P = .400, 2-sided Student t test). AS1842856 decreases viability (C) and induces apoptosis (D) in BCP-ALL cells in a time- and dose-dependent manner. Cells were incubated in the presence of increasing concentrations of AS1842856. The cell number and viability was measured by trypan blue exclusion at indicated time points. Apoptosis was measured using annexin V/PI staining. Data are shown as means of specific cell death ± SD (N = 3). (E) CASP3 cleavage, detection by immunoblot (RS4;11 and 018Z, 40 nM; UoCB6, 80 nM AS1842856), a representative of 3 independent experiments is shown. (F-G) NALM-6 cells were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-wtFOXO1 (WTF1)–expressing vector. For protein analysis, cells were sorted 3 days after transduction (F). (G) Four to 6 days after transduction cells were treated with AS1842856. Percentages of GFP+ cells (by flow cytometry) and the total number of live cells (by cell counting) were measured at indicated time points. Data are shown as fold changes normalized to the initial number of live GFP+ cells. The number of live GFP+ cells was calculated as N × %GFP+ cells/100, where N is the number of live cells per well and %GFP+ is the percentage of GFP+ cells. Data are shown as mean ± SD of 3 independent experiments. ANOVA, *P < .05; **P < .01; ***P < .001, and ****P < .0001, respectively.
To address the specificity of AS1842856, we overexpressed wtFOXO1 in NALM-6 cells (Figure 4F) and exposed them to 40 or 80 nM of AS1842856 assessing proportions of GFP+ and numbers of viable cells. Overexpression of wtFOXO1 did not affect growth of NALM-6 cells compared with control-transduced cells. Incubation of control-transduced cells with AS1842856 led to partial (40 nM) or complete (80 nM) inhibition of cell growth (Figure 4G). Importantly, overexpression of wtFOXO1 completely blocked the cytotoxic effect of AS1842856 at 40 nM and led to a partial rescue of the anti-FOXO effect at the higher concentration of 80 nM (Figure 4G).
AS1842856 represses FOXO1 target genes
To investigate molecular mechanisms of the antileukemic activity of AS1842856, we analyzed gene expression of RS4;11 and UoCB6 cell lines exposed to AS1842856 for 24 hours to identify early transcriptome changes. Eight hundred sixteen probe sets were modulated >1.5-fold in comparison with the vehicle-treated control group (supplemental Table 5). Among the top 20 genes downregulated in both cell lines, 9 known transcriptional targets of FOXO were identified, namely, TCL1A, VPREB3, IL1B, KLHL24, MYC, ID3, CCND3, BNIP3L, and FAIM3 (Figure 5A; supplemental Tables 7 and 8). Moreover, by GSEA we found a significant negative enrichment of FOXO1 associated genes in the “AS1842856” signature (Figure 5B; supplemental Table 6). In addition, by qRT-PCR we validated downregulation of known FOXO1 targets (Figure 5C; supplemental Table 8) including TCL1A and MYC that have an oncogenic function in BCP-ALL,6,40 as well as CCND3, VPREB3, and IL1B, which are highly expressed in pre–B cells. Interestingly, similar to FOXO1 knockdown, AS1842856 downregulated the mTORC2 protein RICTOR and mTORC1 activity (supplemental Figure 5).
AS1842856 represses the FOXO1 signature and downregulates CCND3 in BCP-ALL cell lines. (A-B) RS4;11 and UoCB6 cell lines were incubated with AS1842856 for 24 hours at concentrations corresponding to IC50 values (40 nM and 80 nM, respectively). The raw data were analyzed using the Genesifter software (robust multichip average [RMA] normalization) and genes differentially regulated at least in 1 cell line were filtered (threshold of 1.5; ANOVA; Benjamini and Hochberg correction, adjusted P < .05). (A) Top 20 probe-sets downregulated in both cell lines, direct FOXO targets are shown in red. (B) GSEA. Genes of the FOXO1 activation signature are repressed by AS1842856. Direction of phenotype comparison: “AS1842856 vs control.” Gene signature “FOXO1_UP-BCP-ALL” comprises genes upregulated more than twofold in BCP-ALL cell line RCH-ACV by FOXO1 induction6 (supplemental Table 6). (C) Validation of gene expression profiling. The qRT-PCR data were quantified by the 2−ΔΔCT method; data are shown as mean ± SD, N = 3. Statistical significance was calculated by ANOVA. *P < .05; **P < .01; ***P < .001. (D) CCND3 protein expression in cells incubated with AS1842856 (40 nM). A representative immunoblot of 2 independent experiments is shown. (E-F) CCND3 overexpression protects BCP-ALL cell lines from the cytotoxic effect of AS1842856. BCP-ALL cell lines were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-CCND3 (C3)–expressing vector. For protein analysis, cells were sorted 3 days after transduction (E). After transduction (4-6 days), cells were treated with AS1842856. Percentages of GFP+ and total numbers of live cells were measured at indicated time points. The data are shown as fold change normalized to the initial number of live GFP+ cells. The number of live GFP+ cells was calculated as N × %GFP+ cells/100, where N is the number of live cells per well and %GFP+ is the percentage of GFP+ cells. The data are shown as mean ± SD (N = 3) (F). (G-H) NALM-6 cells transduced with empty vector (EV) or with SF-LV-CCND3 (C3)–expressing vector were sorted 5 days after transduction (day 0) and incubated with 80 nM AS1842856 for another 6 days. Cell lysates were prepared at indicated time points, and protein expression levels were assessed by immunoblot (N = 2) (G). (H) Numbers of viable cells at indicated time points (N = 3). (I-J) Overexpression of CCND3 rescues BCP-ALL cells from cell cycle arrest (I) and apoptosis (J) induced by AS1842856. Cells were harvested on day 3 after start of treatment, cell cycle distribution was analyzed by PI staining (I), and apoptosis was measured using annexin V-PE/7-aminoactinomycin staining (J). Each point represents mean ± SD of 3 independent experiments. ANOVA, ****P < .0001.
AS1842856 represses the FOXO1 signature and downregulates CCND3 in BCP-ALL cell lines. (A-B) RS4;11 and UoCB6 cell lines were incubated with AS1842856 for 24 hours at concentrations corresponding to IC50 values (40 nM and 80 nM, respectively). The raw data were analyzed using the Genesifter software (robust multichip average [RMA] normalization) and genes differentially regulated at least in 1 cell line were filtered (threshold of 1.5; ANOVA; Benjamini and Hochberg correction, adjusted P < .05). (A) Top 20 probe-sets downregulated in both cell lines, direct FOXO targets are shown in red. (B) GSEA. Genes of the FOXO1 activation signature are repressed by AS1842856. Direction of phenotype comparison: “AS1842856 vs control.” Gene signature “FOXO1_UP-BCP-ALL” comprises genes upregulated more than twofold in BCP-ALL cell line RCH-ACV by FOXO1 induction6 (supplemental Table 6). (C) Validation of gene expression profiling. The qRT-PCR data were quantified by the 2−ΔΔCT method; data are shown as mean ± SD, N = 3. Statistical significance was calculated by ANOVA. *P < .05; **P < .01; ***P < .001. (D) CCND3 protein expression in cells incubated with AS1842856 (40 nM). A representative immunoblot of 2 independent experiments is shown. (E-F) CCND3 overexpression protects BCP-ALL cell lines from the cytotoxic effect of AS1842856. BCP-ALL cell lines were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-CCND3 (C3)–expressing vector. For protein analysis, cells were sorted 3 days after transduction (E). After transduction (4-6 days), cells were treated with AS1842856. Percentages of GFP+ and total numbers of live cells were measured at indicated time points. The data are shown as fold change normalized to the initial number of live GFP+ cells. The number of live GFP+ cells was calculated as N × %GFP+ cells/100, where N is the number of live cells per well and %GFP+ is the percentage of GFP+ cells. The data are shown as mean ± SD (N = 3) (F). (G-H) NALM-6 cells transduced with empty vector (EV) or with SF-LV-CCND3 (C3)–expressing vector were sorted 5 days after transduction (day 0) and incubated with 80 nM AS1842856 for another 6 days. Cell lysates were prepared at indicated time points, and protein expression levels were assessed by immunoblot (N = 2) (G). (H) Numbers of viable cells at indicated time points (N = 3). (I-J) Overexpression of CCND3 rescues BCP-ALL cells from cell cycle arrest (I) and apoptosis (J) induced by AS1842856. Cells were harvested on day 3 after start of treatment, cell cycle distribution was analyzed by PI staining (I), and apoptosis was measured using annexin V-PE/7-aminoactinomycin staining (J). Each point represents mean ± SD of 3 independent experiments. ANOVA, ****P < .0001.
Because AS1842856 also inhibited CCND3 protein expression (Figure 5D), we analyzed its role in AS1842856 mediated cytotoxicity. To this end, RS4;11, 018Z, and NALM-6 cell lines overexpressing CCND3 (Figure 5E), were exposed to the inhibitor at toxic concentrations. CCND3 almost completely reversed the cytotoxic effect of AS1842856 in all 3 lines (Figure 5F).
To clarify the mechanism of CCND3-dependent cytoprotection, we sorted CCND3-expressing cells and analyzed phosphorylation of the CCND3-CDK4/6 target RB1 and CDKN1B expression. Overexpression of CCND3 restored AS1842856-induced downregulation of RB1 phosphorylation and decreased the expression of CDKN1B (Figure 5G). This was associated with restoration of cell viability (Figure 5H) and abolished cell cycle arrest (Figure 5I) and apoptosis (Figure 5J).
Thus, we conclude that AS1842856 mimics the effects of FOXO1 knockdown in BCP-ALL and that downregulation of CCND3 contributes to AS1842856 cytotoxicity.
Ex vivo and in vivo antileukemia activity of the FOXO1 inhibitor AS1842856
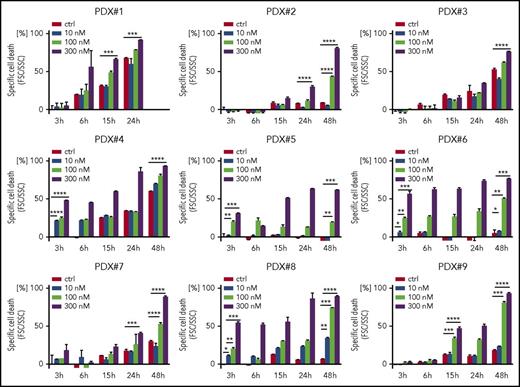
Because we found a high sensitivity of BCP-ALL cells for both genetic and pharmacological FOXO1 depletion, we addressed the cytotoxic activity of AS1842856 on primary, patient-derived leukemia cells. Altogether 9 individual patient-derived leukemia samples isolated from xenografted ALL bearing recipients (supplemental Table 1B) were exposed to increasing concentrations of AS1842856, and leukemia cell survival was assessed over time compared with vehicle-treated cells. The inhibitor showed clear cytotoxic activity in all 9 xenograft leukemias with a dose- and time-dependent increase in cell death (Figure 6).
Antileukemia activity of AS1842856 on primary, PDX ALL. Primary PDX cells were exposed to AS1842856 at concentrations of 10, 100, and 300 nM. Cell death was measured by flow cytometry according to FSC/SSC criteria. Each column represents mean ± SD of 3 independent measurements. A 2-tailed Student t test was used to compare the differences between 2 groups. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Antileukemia activity of AS1842856 on primary, PDX ALL. Primary PDX cells were exposed to AS1842856 at concentrations of 10, 100, and 300 nM. Cell death was measured by flow cytometry according to FSC/SSC criteria. Each column represents mean ± SD of 3 independent measurements. A 2-tailed Student t test was used to compare the differences between 2 groups. *P < .05; **P < .01; ***P < .001; ****P < .0001.
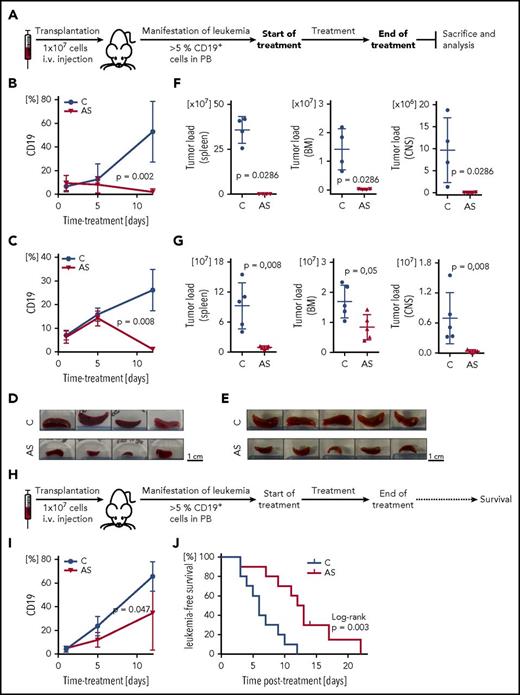
Based on these findings, we investigated the efficacy of FOXO1 inhibition in a preclinical setting in vivo. Two high-risk leukemia xenograft samples (PDX#1, derived from a patient who later on encountered an early relapse, and PDX#7, derived from an infant MLL/ENL rearranged ALL) were transplanted onto recipient animals (Figure 7A). Proportions of human ALL cells (huCD19+ cells) were assessed in the recipient’s peripheral blood clearly showing decreasing percentages of leukemia cells upon AS1842856 administration (50 mg/kg; Figure 7B-C). After therapy, a clearly reduced leukemia burden was observed in the AS1842856-treated group compared with vehicle treated animals with macroscopically less enlarged spleens (Figure 7D-E), and substantially lower leukemia infiltration in spleen, bone marrow, and central nervous system (Figure 7F-G).
Preclinical in vivo antileukemia activity of FOXO1 inhibition by AS1842856. (A) Experimental layout, end point analysis. Recipient animals were transplanted with human BCP-ALL (PDX#1: B, D, F; PDX#7: C, E, G), and upon appearance of human ALL in peripheral blood (≥5% huCD19+ cells), recipients were treated with either AS1842856 (AS, 50 mg/kg) or DMSO (control, C). At the end of therapy after 11 days, animals were euthanized, and leukemia burden was analyzed in different organ compartments showing clearly decreased leukemia cell numbers in peripheral blood (PB) (B-C), reduced spleen size (D-E), and significantly lower leukemia loads in spleen, bone marrow (BM), and central nervous system (CNS) (F-G), compared with control treated animals. PDX#1 n = 4, PDX#7 n = 5 mice per group, Mann-Whitney U test. (H) Experimental layout, survival analysis. Upon manifestation of human ALL (≥5% huCD19+ cells in PB), recipient mice transplanted with PDX#1 were treated with AS1842856 (AS, 30 mg/kg) or DMSO (control, C). At the end of treatment (11 days), mice were followed up and monitored for onset of leukemia-related morbidity indicating leukemia reoccurrence. Reduction of ALL cells in PB (I) and prolonged times without leukemia reoccurrence (J) of recipients treated with AS1842856 compared with control treated recipients. N = 10 per group, Mann-Whitney U test (I) and Kaplan-Meier analysis, log-rank test (J).
Preclinical in vivo antileukemia activity of FOXO1 inhibition by AS1842856. (A) Experimental layout, end point analysis. Recipient animals were transplanted with human BCP-ALL (PDX#1: B, D, F; PDX#7: C, E, G), and upon appearance of human ALL in peripheral blood (≥5% huCD19+ cells), recipients were treated with either AS1842856 (AS, 50 mg/kg) or DMSO (control, C). At the end of therapy after 11 days, animals were euthanized, and leukemia burden was analyzed in different organ compartments showing clearly decreased leukemia cell numbers in peripheral blood (PB) (B-C), reduced spleen size (D-E), and significantly lower leukemia loads in spleen, bone marrow (BM), and central nervous system (CNS) (F-G), compared with control treated animals. PDX#1 n = 4, PDX#7 n = 5 mice per group, Mann-Whitney U test. (H) Experimental layout, survival analysis. Upon manifestation of human ALL (≥5% huCD19+ cells in PB), recipient mice transplanted with PDX#1 were treated with AS1842856 (AS, 30 mg/kg) or DMSO (control, C). At the end of treatment (11 days), mice were followed up and monitored for onset of leukemia-related morbidity indicating leukemia reoccurrence. Reduction of ALL cells in PB (I) and prolonged times without leukemia reoccurrence (J) of recipients treated with AS1842856 compared with control treated recipients. N = 10 per group, Mann-Whitney U test (I) and Kaplan-Meier analysis, log-rank test (J).
Finally, we asked whether the reduced leukemia load upon FOXO1 inhibition would translate into prolonged survival. In an additional experiment, leukemia-bearing recipients (PDX#1, ≥5% human CD19+ cells in PB) were treated with AS1842856 (30 mg/kg) or vehicle for 11 days. After termination of treatment, recipients were followed up for clinical signs of leukemia manifestation and euthanized at onset of leukemia related morbidity estimating times to leukemia reoccurrence for each animal (Figure 7H). In addition, proportions of human ALL cells were measured in the recipient’s blood, showing a delayed growth of human ALL cells (Figure 7I). Importantly, AS1842856-treated animals showed a significantly longer time without leukemia reoccurrence as compared with vehicle-treated mice (Figure 7J). Of note, administration of AS1842856 was well tolerated by the animals.
Thus, inhibition of FOXO1 by a small-molecule inhibitor resulted in a significant antileukemia activity, ex vivo and, importantly, showed clear preclinical in vivo efficacy in 2 high-risk leukemia PDXs.
Discussion
Using in vitro, ex vivo, and in vivo models, we demonstrated an essential role of tightly regulated FOXO1 activity in BCP-ALL. We identified CCND3 downregulation as an important causative factor of cell cycle arrest and apoptosis induced by FOXO1 depletion. Moreover, we showed clear antileukemia activity of pharmacological FOXO1 inhibition, pointing to FOXO1 as a potential therapeutic target.
This finding on an antileukemia effect of FOXO1 repression apparently contradicts the commonly assumed tumor suppressive role of FOXO1 functioning as an ultimate target of essential oncogenic pathways like PI3K-AKT and ERK/MAPK.3,41 However, given that the PI3K-AKT and RAS-ERK pathways are tightly regulated in pre-BCR+ and BCR-ABL1+ BCP-ALL, this might not be surprising.42 Similar to PI3K-AKT and RAS-ERK, activated FOXO1 regulates proliferation and survival of BCP-ALL in accordance with the “Goldilocks principle,” with only intermediate activity favoring tumor maintenance.42 Importantly, this bimodal type of response to FOXO1 activation/inhibition is not restricted to B-cell lineage and has also been described for instance in embryonic endothelial cells.36
We identified an important role of CCND3 downregulation for cell cycle arrest and apoptosis induction upon FOXO1 depletion. Induction of CCND3 upon FOXO1 activation is responsible for pro– and pre–B cell proliferation and block of differentiation.32,43 CCND3 is, in addition to cell cycle regulation, an antiapoptotic factor and genetic or pharmacological inactivation leads to cell death in T-ALL,33,34 Burkitt lymphoma,44 and B-cell chronic lymphocytic leukemia.45 Mechanistically, the CCND3-CDK6 complex phosphorylates enzymes increasing production of reduced NAD phosphate and glutathione.46 Importantly, along with its role in FOXO1 depletion-induced cell death, we showed an essential role of CCND3 for BCP-ALL maintenance and demonstrated the feasibility of BCP-ALL therapy by CDK4/6 inhibitors.
Depletion of FOXO1 also downregulated MYC expression and inhibited of mTORC1 activity. Downregulation of MYC upon FOXO1 knockdown was also described in HUVECs36 and in pre–B ALL after PI3K-AKT activation.17 We found that CCND3 counteracted CDKN1B upregulation induced by FOXO1 knockdown and restored MYC expression. Given that CDKN1B decreases MYC stability,47 it is conceivable that repression of MYC can be, at least partially, explained by CCND3 depletion and therefore plays an auxiliary role.
Knockdown of FOXO1 decreased mTORC1 activity in HUVECs.36 mTORC1 supports cell cycle progression,48,49 and inactivation of either mTORC1 or mTORC2 led to growth arrest and cell death of ALL cell lines and PDXs.50 Our experiments confirmed a role of FOXO1 for mTOR activity in BCP-ALL. To the contrary, protection from the cytotoxic effect of FOXO1 knockdown by CCND3 overexpression was not associated with restoration of mTORC1 activity or RICTOR expression, indicating a redundant role of these factors.
We demonstrated the feasibility of pharmacological FOXO1 inactivation for antitumor therapy using a small-molecular weight inhibitor. It was selected as a compound that selectively binds to FOXO1 protein and represses transactivation of the FOXO1 gluconeogenic targets G6PC and PCK1 to treat type 2 diabetes.39 Importantly, given that genes regulated by FOXO1 are highly cell-type specific,51 functional and molecular consequences of genetic and pharmacological FOXO1 inhibition are also highly conserved in other systems. Both genetic and pharmacological inactivation of FOXO1 stimulated proliferation and protected pulmonary artery smooth muscle cells from apoptosis,52 potentiated regeneration of pancreatic β-cells and restored insulin secretion in diabetic mice,53 repressed autophagy in the HCT116 colon cancer cell line.53 In addition, AS1842856 blocked senescence in ovarian cancer cells,54 protected DLBCL cell lines from FOXO1-dependent doxorubicin-induced cell death,55 and reduced autophagy and cell death in muscle cells by downregulation of FOXO targets atrogin-1 and MuRF-1.56 Importantly, in all these studies AS1842856 stimulated proliferation in normal tissues or protected tumor cells from cell death, although it was used at similar or higher concentrations than in our experiments. Only AML cell lines, which are dependent on FOXO1 expression,57 showed AS1842856 sensitivity.58
Given that FOXO1 is a versatile transcription factor involved in regulation of different biochemical processes in virtually all systems and organs,59 the investigation of possible side effects is important issue. Nevertheless, we did not find reports on severe complications of FOXO1 knockdown or pharmacological depletion. There are 4 FOXO family members, which share partially redundant functions. Interestingly, despite tumor suppressive activity, only deletion of at least 3 FOXOs (1, 3, and 4) leads to formation of tumors in mice. No tumors appeared if one of the FOXOs (including FOXO1) was not deleted.51 Administration of AS1842856 increases number of insulin positive cells in pancreas of diabetic mice, but not in healthy mice.53 AS1842856 normalizes blood glucose levels only in diabetic mice upon treatment with doses of 30 mg/kg; however, in healthy mice AS1842856 does not decrease glucose even at concentrations of 100 mg/kg,39 indicating no risk hypoglycemia in nondiabetic individuals. It seems that tumors, but not normal tissues, are addicted on FOXO1. Thus, knockout of FOXO1 does not influence cell cycle progression and apoptosis in HSCs and myeloid progenitors60 but induced apoptosis in AML cells.57 The sensitivity of CD34+ hematopoietic stem and progenitor cells to AS1842856 was >10-fold higher that of AML preleukemic cells and AML cell lines.58
Most importantly, inhibition of FOXO1 by the small-molecule inhibitor AS1842856 demonstrated a clear antileukemia activity on a series of primary, patient-derived pediatric ALL samples with effective leukemia reduction in hematopoietic and lymphoid organ compartments and particularly in the central nervous system, ultimately leading to prolonged survival in our preclinical in vivo model.
Taken together, we conclude that tightly regulated FOXO1 expression is essential for the maintenance of BCP-ALL. The antileukemic effect of FOXO1 depletion did not depend on the type of oncogenic mutations or differentiation status of the cell lines, pointing to a universal role of FOXO1 in the oncogenic program of BCP-ALL. Our proof-of-concept study warrants additional investigations of AS1842856 to develop a pharmaceutical drug that might be efficient against BCP-ALL. In addition, it might boost a search for new small molecular weight inhibitors targeting FOXO family transcription factors.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE86067).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Kick and S. Volk for excellent technical assistance, E. Kelsch and M. Buck (Institute of Pathology, University of Ulm) for short tandem repeats genotyping of the cell lines, and the Core Facilities “Cell Sorting” and “Genomics,” Ulm University Medical Faculty, Ulm, Germany.
This work was supported in part by grant 110564 from Deutsche Krebshilfe eV (T.W. and A.U.) and by German Research Foundation, SFB 1074, B6 (K.-M.D. and L.H.M.). S.D. and F.G. were supported by the International Graduate School in Molecular Medicine, Ulm, Germany.
Authorship
Contribution: A.U. and T.W. were responsible for conception and design; A.U. and L.H.M. were responsible for development of methodology; F.W., S.D., F.G., C.D.O., K.H., T.M., S.E., S.M.E., and F.S. acquired data; F.W., S.D., K.H., and A.U. performed analysis and interpretation of data; and F.W., S.D., L.H.M., K.-M.D., T.W., and A.U. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexey Ushmorov, Institute of Physiological Chemistry, University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: alexey.ushmorov@uni-ulm.de; and Thomas Wirth, Institute of Physiological Chemistry, University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: thomas.wirth@uni-ulm.de.
References
Author notes
F.W. and S.D. are joint first authors.
T.W., L.H.M., and A.U. are joint last authors.





![Figure 5. AS1842856 represses the FOXO1 signature and downregulates CCND3 in BCP-ALL cell lines. (A-B) RS4;11 and UoCB6 cell lines were incubated with AS1842856 for 24 hours at concentrations corresponding to IC50 values (40 nM and 80 nM, respectively). The raw data were analyzed using the Genesifter software (robust multichip average [RMA] normalization) and genes differentially regulated at least in 1 cell line were filtered (threshold of 1.5; ANOVA; Benjamini and Hochberg correction, adjusted P < .05). (A) Top 20 probe-sets downregulated in both cell lines, direct FOXO targets are shown in red. (B) GSEA. Genes of the FOXO1 activation signature are repressed by AS1842856. Direction of phenotype comparison: “AS1842856 vs control.” Gene signature “FOXO1_UP-BCP-ALL” comprises genes upregulated more than twofold in BCP-ALL cell line RCH-ACV by FOXO1 induction6 (supplemental Table 6). (C) Validation of gene expression profiling. The qRT-PCR data were quantified by the 2−ΔΔCT method; data are shown as mean ± SD, N = 3. Statistical significance was calculated by ANOVA. *P < .05; **P < .01; ***P < .001. (D) CCND3 protein expression in cells incubated with AS1842856 (40 nM). A representative immunoblot of 2 independent experiments is shown. (E-F) CCND3 overexpression protects BCP-ALL cell lines from the cytotoxic effect of AS1842856. BCP-ALL cell lines were transduced with SF-LV-cDNA-eGFP empty vector (EV) or with SF-LV-CCND3 (C3)–expressing vector. For protein analysis, cells were sorted 3 days after transduction (E). After transduction (4-6 days), cells were treated with AS1842856. Percentages of GFP+ and total numbers of live cells were measured at indicated time points. The data are shown as fold change normalized to the initial number of live GFP+ cells. The number of live GFP+ cells was calculated as N × %GFP+ cells/100, where N is the number of live cells per well and %GFP+ is the percentage of GFP+ cells. The data are shown as mean ± SD (N = 3) (F). (G-H) NALM-6 cells transduced with empty vector (EV) or with SF-LV-CCND3 (C3)–expressing vector were sorted 5 days after transduction (day 0) and incubated with 80 nM AS1842856 for another 6 days. Cell lysates were prepared at indicated time points, and protein expression levels were assessed by immunoblot (N = 2) (G). (H) Numbers of viable cells at indicated time points (N = 3). (I-J) Overexpression of CCND3 rescues BCP-ALL cells from cell cycle arrest (I) and apoptosis (J) induced by AS1842856. Cells were harvested on day 3 after start of treatment, cell cycle distribution was analyzed by PI staining (I), and apoptosis was measured using annexin V-PE/7-aminoactinomycin staining (J). Each point represents mean ± SD of 3 independent experiments. ANOVA, ****P < .0001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/26/10.1182_blood-2017-10-813576/4/m_blood813576f5.jpeg?Expires=1768086430&Signature=cjJngkR1w6On54M-aGCN6qoPcF59x0yq1Cpi9r4fxzCTGgJRt4-5tJ4I4ik5Wv5tMNQ9-yoXpAygAiS3X-2C~U~jhKhq6cplZAUNF~7gCcg5fRtywlFSTMm7m0VvjBB-EvtU9Q2F9mwpHT45tPxL9yxaAjm3Jq5g1QIzrPR8gDKWDe8XQpB~YZwh0yPLxUuhhXm2z88OpRYvXCMmNCdLBJWKTWfQMUm7TISArGF9IBIBsK0ZSS8ZnTPOh8tCZjUJLp5W8TFWWRWX9ZkD11RDUlnt-IGEI~~Hb4UOzZ5rlGujfn4b72spqX0yWjmTkNkQqHxWv6XxbPTYw0wr-YtuJg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal