Key Points
CLL cells overexpress a well-defined tumor suppressor Par-4, which promotes malignant B-CLL growth and is regulated through BCR signaling.
Robust regulation of cell-cycle modulator p21/WAF1 by Par-4 in CLL cells.
Abstract
Prostate apoptosis response-4 (Par-4), a proapoptotic tumor suppressor protein, is downregulated in many cancers including renal cell carcinoma, glioblastoma, endometrial, and breast cancer. Par-4 induces apoptosis selectively in various types of cancer cells but not normal cells. We found that chronic lymphocytic leukemia (CLL) cells from human patients and from Eµ-Tcl1 mice constitutively express Par-4 in greater amounts than normal B-1 or B-2 cells. Interestingly, knockdown of Par-4 in human CLL-derived Mec-1 cells results in a robust increase in p21/WAF1 expression and decreased growth due to delayed G1-to-S cell-cycle transition. Lack of Par-4 also increased the expression of p21 and delayed CLL growth in Eμ-Tcl1 mice. Par-4 expression in CLL cells required constitutively active B-cell receptor (BCR) signaling, as inhibition of BCR signaling with US Food and Drug Administration (FDA)–approved drugs caused a decrease in Par-4 messenger RNA and protein, and an increase in apoptosis. In particular, activities of Lyn, a Src family kinase, spleen tyrosine kinase, and Bruton tyrosine kinase are required for Par-4 expression in CLL cells, suggesting a novel regulation of Par-4 through BCR signaling. Together, these results suggest that Par-4 may play a novel progrowth rather than proapoptotic role in CLL and could be targeted to enhance the therapeutic effects of BCR-signaling inhibitors.
Introduction
Chronic lymphocytic leukemia (CLL) is defined by accumulation of clonally expanded CD5+ and CD19+ B lymphocytes in blood, bone marrow, and secondary lymphoid organs with impaired apoptotic mechanisms.1-4 CLL can be classified into mutated (M-CLL) and unmutated (U-CLL) forms based on the extent of mutation in the B-cell receptor (BCR) variable region genes. The U-CLL group exhibits increased BCR signaling, more aggressive disease, and worse prognosis.2 Recently, BCR signaling in CLL has become one of the most promising therapeutic targets after successful clinical trials with multiple kinase inhibitors,3 as it is required for the survival of malignant B cells and is constitutively activated in CLL.3,4 Small molecule inhibitors toward Bruton tyrosine kinase (Btk) and phosphatidylinositol-3 kinase (PI3K) have led to higher overall response rates in patients, but complete responses are rare, necessitating further studies.5
Prostate apoptosis response-4 (Par-4) is a tumor suppressor that is downregulated by promoter methylation in ∼30% of endometrial cancers and acute lymphoblastic leukemia.6,7 It was originally identified by its upregulation during apoptosis of prostate cancer cells and later shown to have a cancer-selective mechanism of inducing cell death through a specific selective for apoptosis of cancer (SAC) domain.8 Par-4 is activated by protein kinase A (PKA) phosphorylation allowing Par-4 to translocate to the nucleus and inhibit NF-κB activity in cancer cells but not normal cells.9 Par-4 directly interacts with Wilms tumor-1 protein,10 and downregulates Bcl-2 in nonhematopoietic cells.11,12 Par-4 is also secreted from cells and induces apoptosis by activating the Fas-signaling pathway through binding to the extracellular receptor, GRP78.13 A study by Chow et al suggested that CLL patients who express high levels of Par-4 respond better to imatinib treatment.14 A 2011 study found increased Par-4 expression in CD38+ CLL subgroup and advanced-stage patients, although such a correlation was not found in another study.12,15
Here, we sought to understand the role of Par-4 in CLL and the regulatory mechanisms underlying its expression in CLL using the Eµ-Tcl1 mouse model. This mouse develops a CLL-like disease by 9 to 13 months of age, due to a B-cell–specific overexpression of the oncogene, T-cell leukemia 1 (Tcl1).16 Elevated Tcl1 expression is associated with the more aggressive forms of CLL in human patients.17 Surprisingly, we observed that CLL cells express more Par-4 protein and messenger RNA (mRNA) than all normal B-cell subsets studied. Similarly, human CLL samples have elevated levels of Par-4 protein compared with normal donors. Because CLL cell survival requires tonic BCR signaling,18 we hypothesized that BCR signaling may regulate constitutive Par-4 expression in CLL cells. Our studies support this hypothesis and show that Par-4 plays a novel progrowth rather than a proapoptotic role in CLL.
Materials and methods
CLL patients
Human patients with CLL were recruited from the Hematology and Oncology Clinics at the University of Kentucky (UK) and The Ohio State University (OSU). The diagnosis of CLL was confirmed by board-certified hematologists. Each patient gave informed consent, approved through the UK or OSU Institutional Review Boards in accordance with the Declaration of Helsinki. UK patient information was confirmed through a linkage with the Kentucky Cancer Registry performed by the Markey Cancer Center (MCC) Cancer Research Informatics Shared Resource Facility. Peripheral blood CLL cells were purified through Ficoll-Paque density gradients and confirmed for CLL phenotype by CD45+CD5+CD19+ staining via flow cytometry. Healthy control donors were obtained from leukopak units purchased from the Kentucky Blood Bank (Lexington, KY) after quality testing. Human cells were cultured in complete RPMI 1640 media.
Mice
Eμ-Tcl1 and Par-4−/− mice were described previously and were bred in-house.16,19,20 C57BL/6J and NOD-scid IL2rgnullSCF/GM-CSF/IL3 (NSGS) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred in-house. NOD scid IL2rgnull (NSG) mice were obtained from The Jackson Laboratory. Par-4−/−Tcl1 mice were generated by crossing Par-4−/− mice to Eμ-Tcl1 mice, both of which were of C57BL/6J background. Adoptive transfer models were carried out by injecting C57BL/6J mice with 10 × 106 Ficoll-Paque purified splenic Eμ-Tcl1 CLL cells IV via retro-orbital route. CLL development was monitored through submandibular vein bleeding.21 When the percentage of CD5+CD19+ cells in the peripheral blood was >70%, Eμ-Tcl1 mice were carefully monitored for moribund body conditions and euthanized accordingly. Cells were isolated from the spleen of Eμ-Tcl1 mice and confirmed for the CLL phenotype (CD5+CD19+) by flow cytometry (supplemental Figure 1A, available on the Blood Web site).
Cell survival and proliferation assays
Cell survival and proliferation were determined by the 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide (MTT) assay.22 Primary mouse and human CLL cells (2 × 105 cells per well) or Mec-1/OSU-CLL cells (2 × 104 cells per well) were cultured in 96-well flat-bottom plates in 200 µL. The cells were treated with 0 to 40 µM dasatinib, fostamatinib, or ibrutinib dissolved in dimethyl sulfoxide (DMSO) for 48 hours and incubated with 0.5 mg/mL MTT for 4 hours followed by solubilization and spectrophotometric measurements at 560 nm and 690 nm. DMSO concentration did not exceed 0.02% of culture medium and had no measurable effect on cell viability.
shRNA lentiviral infection
Mec-1 CLL cells were transduced with lentiviruses expressing Lyn short hairpin RNA (shRNA) and Par-4 shRNA. Lyn shRNA gene sets were purchased through ThermoFisher Scientific. Par-4 shRNA constructs were expressed in the pLKO.1 lentiviral vector. Lentiviral particles from the supernatants were transduced into Mec-1 cells in complete Iscove modified Dulbecco's medium with 10 μg/mL Polybrene by centrifugation for 90 minutes at 2800 rpm at 10°C. After 24 hours at 37°C, cells were transferred to fresh media and selected by puromycin resistance. Gene-silencing efficiency was analyzed by immunoblotting for respective proteins. Control shRNA and Par-4 shRNA Mec-1 cell growth rates were monitored by counting total cell numbers at each passage. Individual control and Par-4 shRNA-expressing clones were isolated by limiting dilution in the presence of puromycin.
In vivo tumor study
Mec-1 cells (2 × 106) expressing Par-4 shRNA or control shRNA were injected subcutaneously into the right or left flanks of 6 NSGS mice, respectively, with a 1:1 ratio of Matrigel in a 100-μL volume. Tumor size was measured using electronic caliper and tumor volumes were calculated by length (in millimeters) by width-squared (in millimeters). Prior to injection, Par-4 knockdown was verified by immunoblot analysis.
Additional materials and methods can be found in supplemental Materials and methods.
Results
Eμ-Tcl1 CLL cells have elevated expression of Par-4
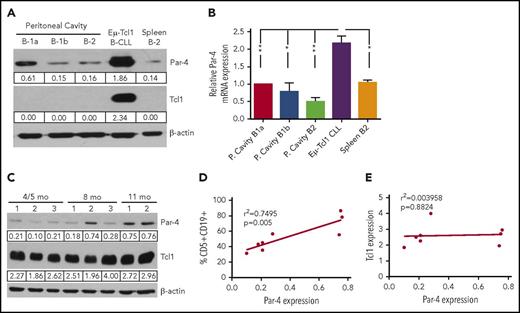
Constitutive Par-4 expression has been described in CLL cells, but most previous studies compared CLL cells to peripheral blood B cells.12,15 Whole-blood B cells are mostly made of the B-2 cell subset, but B-1a cells are the likely cells of origin for U-CLL and Eμ-Tcl1 CLL.23,24 Hence, we isolated CD5+CD19+ CLL cells from the spleens of Eμ-Tcl1 mice and B-cell subsets from the peritoneum and spleen of C57/BL6J wild-type (WT) mice by fluorescence-activated cell sorting into specific populations of B-1a (CD19+CD5+CD11b+), B-1b (CD19+CD5−CD11b+), and B-2 (CD19+CD5−CD11b−) cells (supplemental Figure 1B).25 Immunoblot and polymerase chain reaction (PCR) analysis indicated that Eμ-Tcl1 CLL cells expressed higher levels of Par-4 protein and mRNA than all of the normal B-cell subsets (Figure 1A-B). B-1a cells expressed more Par-4 compared with the other B-cell populations but only ∼33% of the observed levels in CLL. Because Eμ-Tcl1 CLL cells overexpress the oncogene Tcl1, we questioned whether the increased Par-4 expression in CLL cells was associated with Tcl1 levels. WT B-1a with no detectable Tcl1 expressed more Par-4 than Eμ-Tcl1 B-2 cells with measurable Tcl1 (supplemental Figure 1C). Despite substantial Tcl1 expression by all Eμ-Tcl1 B-cell subsets, Par-4 protein levels were not as high as spleen cells from Eμ-Tcl1 mice with >90% CD5+CD19+ CLL cells. CD5+CD19+ B-cell percentage in the peripheral blood and spleen increased with the age of Eμ-Tcl1 mice26 and immunoblot analysis of whole spleen cells from Eμ-Tcl1 mice of different ages indicated that Par-4 protein expression increased with CLL burden (Figure 1C). There was a positive association between leukemic burden and Par-4 (r2 = 0.7495; P < .0055) (Figure 1D) but not with levels of Tcl1 when both proteins were normalized to β-actin (r2 = 0.003958; P = .8824) (Figure 1E).
Eμ-Tcl1 CLL spleen cells have elevated levels of Par-4 expression compared with normal B-cell subsets, which increases with progression of disease. (A) CLL cells were harvested from the spleen of Eμ-Tcl1 mice. B-cell subsets from C57BL/6J WT mice were isolated from the peritoneal cavity and spleen. Protein lysates of WT mice represent a pool of 20 mice. Levels of Par-4 and Tcl1 were quantified by western blot and normalized to β-actin. (B) Histogram shows relative Par-4 mRNA expression in Eμ-Tcl1 CLL cells and WT B-cell subsets quantified by qRT-PCR. Par-4 mRNA levels were normalized to mouse 18S mRNA expression and were then normalized to B-1a Par-4 mRNA expression. For panels A and B, results are representative of 2 experiments. Error bars represent standard error of the mean (SEM). *P < .05, **P < .001 as determined by the Student t test. (C) Par-4 protein expression increases with age and disease development in spleen cells isolated from the Eμ-Tcl1 mouse. Par-4 and Tcl1 protein expression is normalized to β-actin. Graphs show (D) correlation of Par-4 protein expression and percentage of CD5+CD19+ cells; (E) correlation of Par-4 protein expression and Tcl1 expression in the spleens of mice at specific ages. Each point represents an average of 3 mice. r2 value determined by Pearson coefficient test and P = .0055 (D) and P = .8824 (E) determined by linear regression analysis. P. Cavity, peritoneal cavity.
Eμ-Tcl1 CLL spleen cells have elevated levels of Par-4 expression compared with normal B-cell subsets, which increases with progression of disease. (A) CLL cells were harvested from the spleen of Eμ-Tcl1 mice. B-cell subsets from C57BL/6J WT mice were isolated from the peritoneal cavity and spleen. Protein lysates of WT mice represent a pool of 20 mice. Levels of Par-4 and Tcl1 were quantified by western blot and normalized to β-actin. (B) Histogram shows relative Par-4 mRNA expression in Eμ-Tcl1 CLL cells and WT B-cell subsets quantified by qRT-PCR. Par-4 mRNA levels were normalized to mouse 18S mRNA expression and were then normalized to B-1a Par-4 mRNA expression. For panels A and B, results are representative of 2 experiments. Error bars represent standard error of the mean (SEM). *P < .05, **P < .001 as determined by the Student t test. (C) Par-4 protein expression increases with age and disease development in spleen cells isolated from the Eμ-Tcl1 mouse. Par-4 and Tcl1 protein expression is normalized to β-actin. Graphs show (D) correlation of Par-4 protein expression and percentage of CD5+CD19+ cells; (E) correlation of Par-4 protein expression and Tcl1 expression in the spleens of mice at specific ages. Each point represents an average of 3 mice. r2 value determined by Pearson coefficient test and P = .0055 (D) and P = .8824 (E) determined by linear regression analysis. P. Cavity, peritoneal cavity.
CLL cells secreted high amounts of Par-4 compared with normal and stimulated spleen B cells (supplemental Figure 2A). Secreted Par-4 from CLL cells induced apoptosis of H460 lung cancer cells, which was reduced by inclusion of anti–Par-4 antibody but not control immunoglobulin G (IgG) (supplemental Figure 2B). However, primary human CLL cells were not sensitive to treatment with recombinant Par-4 or the SAC domain, although they were all killed by OSU-2S, an FTY720 derivative (supplemental Figure 2C). Extracellular Par-4 requires the receptor GRP78 to bind and induce extrinsic apoptosis.27 CLL cells have lower levels of basal GRP78 protein compared with other cell lines and this may be one reason why CLL cells were not sensitive to Par-4–mediated apoptosis (supplemental Figure 2D). Because intracellular Par-4 has been shown to function by translocation into nucleus, we quantified Par-4 levels in the cytoplasm and nucleus. CLL samples from different individual Eμ-Tcl1 mice expressed Par-4 both in the cytoplasm and the nucleus (supplemental Figure 2E). Additionally, we adoptively transferred Eμ-Tcl1 CLL cells into syngeneic C57BL/6J (WT) and Par-4−/− mice19 and detected similar levels of Par-4 in the plasma as well as spleens of WT and Par-4−/− mice 4 weeks after transfer (supplemental Figure 3). Thus, Par-4 expressed by CLL cells is secreted in vitro and in vivo, is cytotoxic to cell lines from solid tumors as shown previously, and is able to translocate to the nucleus, which is needed for intracellular Par-4 to induce apoptosis.27 Despite the expression of functional Par-4, CLL cells develop de novo in Par-4+/+Tcl1 mice or grow unimpeded during adoptive transfer into WT or Par-4−/− recipients.
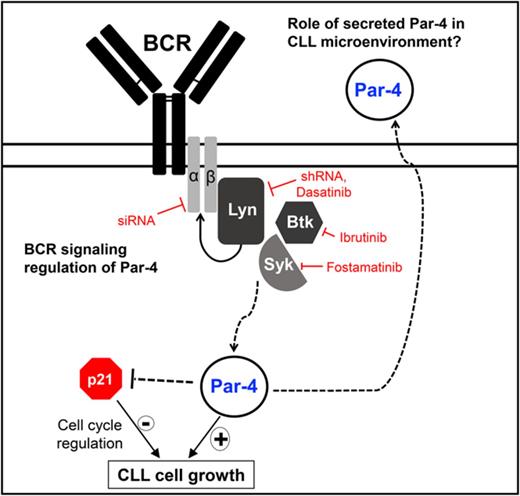
Novel regulation of Par-4 through BCR signaling in CLL
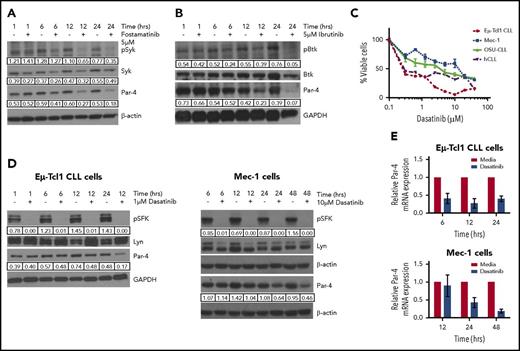
We tested the hypothesis that constitutively elevated expression of Par-4 may be regulated by the well-known tonic BCR signaling in CLL. Accordingly, Eµ-Tcl1 cells have increased phosphorylated Src family kinase (p-SFK) compared with both B-2 as well as B-1 cell subsets (supplemental Figure 1C). Inhibition of Syk or Btk with fostamatinib and ibrutinib, respectively, led to a decrease in Par-4 protein accompanied by a decrease in p-Syk and p-Btk and a decrease in viability, demonstrating the effectiveness of the inhibitors in blocking their target kinases (Figure 2A-B; supplemental Figure 4). In addition to the primary Eμ-Tcl1 CLL cells, we tested the effect of dasatinib on 2 human-derived CLL cell lines, Mec-1 and OSU-CLL,28,29 as well as primary human peripheral blood CLL samples. Similar to the other kinase inhibitors, dasatinib reduced the survival of all CLL cell models in a dose-dependent manner (Figure 2C) and decreased p-SFK without affecting levels of total Lyn kinase (Figure 2D). Reduced SFK activity in Eµ-Tcl1 and Mec-1 CLL cells was accompanied by a decrease in Par-4 protein and mRNA expression (Figure 2D-E), suggesting regulation at the transcript level. Minimal changes in Syk, Btk, and Lyn total protein levels and the changes in Par-4 protein and transcript levels detected at early time points make it unlikely that decrease in Par-4 is due to death of CLL cells after kinase inhibitor treatment. Due to the potential off-target effects of kinase inhibitors, we also used lentiviral-mediated shRNA to target Lyn, as it is required for BCR signaling and survival of CLL cells.30-32 A 50% knockdown of Lyn expression in Mec-1 cells led to reduced Par-4 protein and mRNA expression (Figure 3A). Similar results were obtained with the B-cell lymphoma cell line, LY-3, confirming that inhibition of Lyn leads to a decrease in Par-4 expression (supplemental Figure 5).
Eμ-Tcl1 CLL cells have constitutively activated BCR signaling that regulates Par-4 expression. (A) Western blot of Eμ-Tcl1 CLL cells treated with 5 μM fostamatinib for different time points. (B) Western blot of Eμ-Tcl1 CLL cells treated with 5 μM ibrutinib. (C) Survival of Eμ-Tcl1 CLL cells, 2 human CLL (hCLL) cell lines (Mec-1, OSU-CLL), and primary human CLL cells is dependent on BCR signaling. Cells were treated with different concentrations of dasatinib for 48 hours and viability was measured by MTT assay in triplicates. (D) Primary Eμ-Tcl1 CLL cells (left) or Mec-1 CLL cells (right) were treated with 1 μM dasatinib and the protein lysates were analyzed by western blot. (E) Treatment of Eμ-Tcl1 CLL cells (left) or Mec-1 cells (right) with dasatinib decreases Par-4 mRNA as measured by qRT-PCR. Par-4 mRNA is normalized to mouse 18S mRNA expression. In immunoblots, phosphorylated protein values were normalized to total protein. β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize total protein levels (Lyn, Syk, Btk, and Par-4) in the samples. Results are representative of 3 or more experiments.
Eμ-Tcl1 CLL cells have constitutively activated BCR signaling that regulates Par-4 expression. (A) Western blot of Eμ-Tcl1 CLL cells treated with 5 μM fostamatinib for different time points. (B) Western blot of Eμ-Tcl1 CLL cells treated with 5 μM ibrutinib. (C) Survival of Eμ-Tcl1 CLL cells, 2 human CLL (hCLL) cell lines (Mec-1, OSU-CLL), and primary human CLL cells is dependent on BCR signaling. Cells were treated with different concentrations of dasatinib for 48 hours and viability was measured by MTT assay in triplicates. (D) Primary Eμ-Tcl1 CLL cells (left) or Mec-1 CLL cells (right) were treated with 1 μM dasatinib and the protein lysates were analyzed by western blot. (E) Treatment of Eμ-Tcl1 CLL cells (left) or Mec-1 cells (right) with dasatinib decreases Par-4 mRNA as measured by qRT-PCR. Par-4 mRNA is normalized to mouse 18S mRNA expression. In immunoblots, phosphorylated protein values were normalized to total protein. β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize total protein levels (Lyn, Syk, Btk, and Par-4) in the samples. Results are representative of 3 or more experiments.
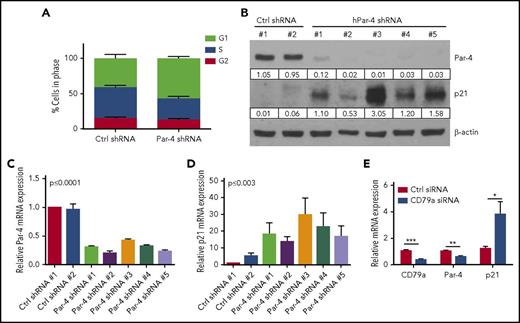
Inhibition and silencing of BCR signaling results in a decrease in Par-4 expression. (A) Mec-1 CLL cells were treated with control (Ctrl) or Lyn-specific shRNA lentivirus and selected through puromycin treatment. Cells were collected at day 17 and total protein and RNA was isolated. Left panel, Levels of Lyn protein. Right panel, Levels of Par-4 mRNA. (B) Human CLL cells were electroporated with CD79a siRNA and incubated for 24 hours. Total protein was isolated and probed for CD79a and Par-4. Western blot represents data from 1 CLL donor with average results for 4 other donors shown as histogram. Ctrl siRNA, n = 2; CD79a siRNA, n = 2. Error bars represent SEM. (C) Surface IgM levels were measured on human CLL cells electroporated with control and CD79a siRNA by flow cytometry (left, representative histogram for 1 patient; middle, average mean fluorescent intensity [MFI] of IgM expression for 4 donors). Total RNA was isolated from cells, and CD79a and Par-4 mRNA were measured by qRT-PCR. Expression values were normalized to human 18S expression performed in triplicate for 4 different donors. *P ≤ .05, **P ≤ .01, ***P ≤ .001 determined by the Student t test.
Inhibition and silencing of BCR signaling results in a decrease in Par-4 expression. (A) Mec-1 CLL cells were treated with control (Ctrl) or Lyn-specific shRNA lentivirus and selected through puromycin treatment. Cells were collected at day 17 and total protein and RNA was isolated. Left panel, Levels of Lyn protein. Right panel, Levels of Par-4 mRNA. (B) Human CLL cells were electroporated with CD79a siRNA and incubated for 24 hours. Total protein was isolated and probed for CD79a and Par-4. Western blot represents data from 1 CLL donor with average results for 4 other donors shown as histogram. Ctrl siRNA, n = 2; CD79a siRNA, n = 2. Error bars represent SEM. (C) Surface IgM levels were measured on human CLL cells electroporated with control and CD79a siRNA by flow cytometry (left, representative histogram for 1 patient; middle, average mean fluorescent intensity [MFI] of IgM expression for 4 donors). Total RNA was isolated from cells, and CD79a and Par-4 mRNA were measured by qRT-PCR. Expression values were normalized to human 18S expression performed in triplicate for 4 different donors. *P ≤ .05, **P ≤ .01, ***P ≤ .001 determined by the Student t test.
To further establish the requirement for BCR, we used small-interfering RNA (siRNA) to target CD79a (Igα) and tested the effect on Par-4 expression in primary human CLL cells. Inhibiting CD79a expression reduces surface BCR expression and the downstream activation of nonreceptor tyrosine kinases required for BCR signaling, eliminating any pleiotropic effects of kinase inhibition.4 siRNA transfection efficiency was ∼47% to 90% (supplemental Figure 6). We observed a decrease in both CD79a and Par-4 expression by human CLL cells with CD79a silencing, further confirming a novel regulation of Par-4 through BCR signaling (Figure 3B). We verified that silencing of CD79a reduced IgM expression levels by flow cytometry, leading to reduced Par-4 mRNA expression in multiple human CLL cell samples (Figure 3C). Extracellular signal-regulated kinase (ERK) signaling is known to be critical for B-cell survival and proliferation downstream of the BCR activation.33-35 Inhibition of ERK1/2 with SCH772984, a well-characterized ERK2 inhibitor, led to a decrease in Par-4 protein expression as well as a decrease in ERK phosphorylation (supplemental Figure 7).36 Interestingly, there was no decrease in Par-4 expression in PC-3 cells after ERK inhibition (supplemental Figure 7), suggesting that Par-4 is downstream of a survival and proliferation pathway important in malignant B cells but not in prostate cancer cells.
Par-4 regulates the growth of CLL cells in vitro and in vivo
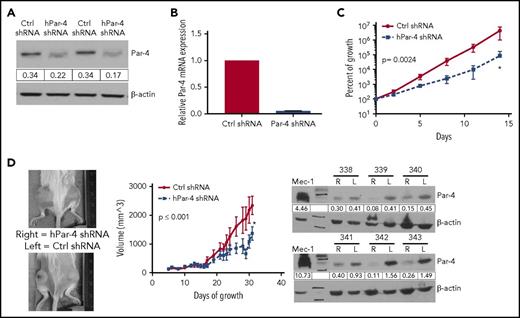
We evaluated the role of Par-4 in CLL by knocking down Par-4 in 2 CLL cell lines. There was a decrease in Par-4 protein and mRNA expression after lentiviral shRNA-mediated knockdown in Mec-1 cells (Figure 4A-B), which led to a decrease in overall growth rate in vitro (Figure 4C; P = .0024). Par-4 knockdown with shRNA led to reduced growth and thymidine incorporation in the OSU-CLL cell line compared with control shRNA treatment (supplemental Figure 8A-C). Par-4 is known to inhibit Akt phosphorylation and reduce Bcl-2 levels,12,37 which were increased in Par-4 knockdown Mec-1 cells confirming specific Par-4 knockdown (supplemental Figure 9). Subcutaneous injection of control shRNA and Par-4 shRNA knockdown Mec-1 cells into left or right flanks of NSGS mice, respectively, induced tumors that were followed for 3 weeks. Par-4 knockdown Mec-1 cells grew more slowly in vivo, mimicking the in vitro results (Figure 4D left 2 panels). Injected tumors retained their knockdown of Par-4 expression in vivo as shown through immunoblot analysis (Figure 4D right panel). Additionally, NSG mice injected with Par-4 shRNA-expressing OSU-CLL cells exhibited improved survival time and delayed growth compared with mice injected with OSU-CLL cells treated with control shRNA (supplemental Figure 8D).
Par-4 knockdown in Mec-1 CLL cells results in reduced growth in vitro and in vivo. (A) Western blot showing a reduction in Par-4 in Mec-1 cells expressing Par-4–specific shRNA. Par-4 protein values were normalized to β-actin. (B) Par-4 mRNA is decreased in Mec-1 cells expressing Par-4 shRNA compared with those expressing a control shRNA. Par-4 mRNA expression was normalized to human 18S and was measured in triplicate. (C) Growth curve of Mec-1 cells expressing control or Par-4–specific shRNA. Curves represent an average of 4 control and 4 Par-4 shRNA-treated clones. Slopes of the curves are different (P = .0024) as calculated by linear regression analysis. (D) Mec-1 cells (2 × 106) treated with control or Par-4–specific shRNA lentivirus were engrafted into NSGS mice (n = 6) subcutaneously with Matrigel. Images represent 2 mice of 6 after 35 days of growth (left). Tumor volumes are plotted as a function of time (middle panel). Tumor volumes were calculated by measuring length and width with a caliper. Difference between the slopes of the 2 lines was found to be statistically significant (P ≤ .001) by linear regression analysis. After 35 days of growth, the tumors were excised and western blot analysis was performed to confirm tumors retained Par-4 knockdown in each mouse throughout the experiment (right panel). The numbers above blots refer to individual mice. Error bars represent SEM. L, left; R, right.
Par-4 knockdown in Mec-1 CLL cells results in reduced growth in vitro and in vivo. (A) Western blot showing a reduction in Par-4 in Mec-1 cells expressing Par-4–specific shRNA. Par-4 protein values were normalized to β-actin. (B) Par-4 mRNA is decreased in Mec-1 cells expressing Par-4 shRNA compared with those expressing a control shRNA. Par-4 mRNA expression was normalized to human 18S and was measured in triplicate. (C) Growth curve of Mec-1 cells expressing control or Par-4–specific shRNA. Curves represent an average of 4 control and 4 Par-4 shRNA-treated clones. Slopes of the curves are different (P = .0024) as calculated by linear regression analysis. (D) Mec-1 cells (2 × 106) treated with control or Par-4–specific shRNA lentivirus were engrafted into NSGS mice (n = 6) subcutaneously with Matrigel. Images represent 2 mice of 6 after 35 days of growth (left). Tumor volumes are plotted as a function of time (middle panel). Tumor volumes were calculated by measuring length and width with a caliper. Difference between the slopes of the 2 lines was found to be statistically significant (P ≤ .001) by linear regression analysis. After 35 days of growth, the tumors were excised and western blot analysis was performed to confirm tumors retained Par-4 knockdown in each mouse throughout the experiment (right panel). The numbers above blots refer to individual mice. Error bars represent SEM. L, left; R, right.
The unexpected result of Par-4 knockdown reducing the growth rate of Mec-1 and OSU-CLL cells (Figure 4; supplemental Figure 8) and increasing the expression of known prosurvival and antiapoptotic proteins (pAkt and Bcl-2; supplemental Figure 9), led us to investigate whether knockdown of Par-4 affected the cell cycle in CLL cells. Using 5 different Par-4 shRNA and 4 control shRNA-expressing clones, we showed that Par-4 knockdown Mec-1 cells had fewer cells entering S phase but more cells in G1 phase, suggesting a reduction in the G1-to-S transition (Figure 5A). Analysis of cell-cycle regulatory proteins showed no change in Cdk1 expression with only modest changes in Cdk2 and Cdk4 in Mec-1 cells upon knocking down Par-4 (supplemental Figure 10). Expression of p21 was increased in the Par-4 knockdown clones compared with clones generated with control shRNA (Figure 5B). Quantitative reverse transcription PCR (qRT-PCR) confirmed the protein expression results with a decrease in Par-4 mRNA and a reciprocal increase in p21 mRNA expression in all 5 Par-4 knockdown clones of Mec-1 cells (Figure 5C-D). Additionally, using human CLL cells, we confirmed that BCR-signaling inhibition through CD79a silencing resulted in a decrease in Par-4 and a reciprocal increase in p21 mRNA (Figure 5E).
Par-4 knockdown results in G1 arrest and increased p21 expression. (A) Mec-1 cell clones expressing control or Par-4–specific shRNA were stained with propidium iodide. Cell-cycle analysis was performed by flow cytometry. Histograms represent mean ± SE of 4 control shRNA clones and 5 Par-4 shRNA clones (G1, P ≤ .0001; S, P ≤ .0002; G2, P = .15 comparing control to Par-4 shRNA-expressing clones). (B) shRNA lentivirus-infected cells were collected and total protein was isolated. Par-4 and p21 proteins were measured through immunoblot analysis. Protein quantifications were normalized to β-actin. (C-D) RNA was isolated from shRNA lentivirus-infected cells. Par-4 mRNA (C) and p21 mRNA (D) were quantified by qRT-PCR and were normalized to human 18S expression (C, P ≤ .0001; D, P ≤ .003 comparing control shRNA clones to Par-4 shRNA clones). (E) Human CLL cells were electroporated with CD79a siRNA and incubated for 72 hours. CD79a, Par-4, and p21 mRNA levels were quantified by qRT-PCR and were normalized to human 18S. Results represent mean ± SEM of triplicate determinations and an average of 4 CLL patient donors.
Par-4 knockdown results in G1 arrest and increased p21 expression. (A) Mec-1 cell clones expressing control or Par-4–specific shRNA were stained with propidium iodide. Cell-cycle analysis was performed by flow cytometry. Histograms represent mean ± SE of 4 control shRNA clones and 5 Par-4 shRNA clones (G1, P ≤ .0001; S, P ≤ .0002; G2, P = .15 comparing control to Par-4 shRNA-expressing clones). (B) shRNA lentivirus-infected cells were collected and total protein was isolated. Par-4 and p21 proteins were measured through immunoblot analysis. Protein quantifications were normalized to β-actin. (C-D) RNA was isolated from shRNA lentivirus-infected cells. Par-4 mRNA (C) and p21 mRNA (D) were quantified by qRT-PCR and were normalized to human 18S expression (C, P ≤ .0001; D, P ≤ .003 comparing control shRNA clones to Par-4 shRNA clones). (E) Human CLL cells were electroporated with CD79a siRNA and incubated for 72 hours. CD79a, Par-4, and p21 mRNA levels were quantified by qRT-PCR and were normalized to human 18S. Results represent mean ± SEM of triplicate determinations and an average of 4 CLL patient donors.
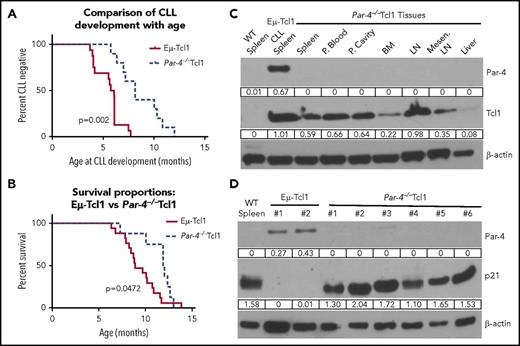
To determine the effect of Par-4 in the development of CLL, Eμ-Tcl1 mice were crossed with Par-4−/− mice and monitored for CLL progression by measuring the CD5+CD19+ cells in the blood. Mice were considered to have CLL if there was a distinct population of 15% to 20% CD5+CD19+ cells in the blood. Figure 6A shows that CLL development is significantly delayed in Par-4−/−Tcl1 mice compared with the Par-4+/+Tcl1 mice (P < .0003). Figure 6B represents the survival analysis for Par-4+/+Tcl1 and Par-4−/−Tcl1 mice. The average age of death of Par-4+/+Tcl1 was 8.9 months vs the 11.97 months for Par-4−/−Tcl1 mice (P < .05). Immunoblot analysis of several tissues from the Par-4−/−Tcl1 mice confirmed deficiency in Par-4, but positive expression of Tcl1 (Figure 6C). Par-4+/+Tcl1 mice expressed Par-4 and Tcl1 in all tissues tested (supplemental Figure 11). Par-4−/−Tcl1 spleen cells expressed higher levels of p21 protein compared with Par-4+/+Tcl1 spleen cells providing in vivo confirmation of the p21 upregulation observed in vitro using Par-4 knockdown cell lines (Figure 6D). For p21 to execute its function to block G1-to-S transition, p21 has to translocate to the nucleus.38 Accordingly, Par-4−/−Tcl1 cells showed a robust increase of p21 in the nucleus compared with Par-4+/+Tcl1 cells, confirming that knockout of Par-4 led to increased expression of functional p21 (supplemental Figure 12).
Loss of Par-4 in the Eμ-Tcl1–transgenic mice delays CLL growth and increases survival. Leukemic status in Par-4+/+ and Par-4−/− EµTcl1 mice was measured by staining of peripheral blood lymphocytes for CD5+CD19+ cells. Animals died of natural progression of disease or were euthanized due to poor body conditions for humane reasons. (A) Percentage of CLL cases detected with age of Par-4+/+Tcl1 and Par-4−/−Tcl1 cohorts over time. P = .0002 comparing the 2 curves by log-rank test (n = 16, Par-4+/+Eμ-Tcl1; n = 10, Par-4−/−Tcl1). (B) Effect of Par-4 loss on the survival of Eµ-Tcl1 mice. Survival curve represents a total of 17 Par-4+/+ Tcl1 mice and 9 Par-4−/−Tcl1 mice. P = .0472 comparing the 2 curves by log-rank test. (C) Tissues from Par-4−/−Tcl1 mice were harvested and expression of Par-4 and Tcl1 proteins was determined by western blot analysis. Par-4 was detected in the spleen of Par-4+/+Tcl1 mouse, but was not present in any of the tissues of the Par-4−/−Tcl1 mouse. (D) Spleens from multiple Eμ-Tcl1 mice and Par-4−/−Tcl1 mice were harvested and total protein was isolated. Immunoblots were probed for Par-4 and p21. Protein expression was normalized to β-actin. LN, lymph node; Mesen. LN, mesenteric lymph node.
Loss of Par-4 in the Eμ-Tcl1–transgenic mice delays CLL growth and increases survival. Leukemic status in Par-4+/+ and Par-4−/− EµTcl1 mice was measured by staining of peripheral blood lymphocytes for CD5+CD19+ cells. Animals died of natural progression of disease or were euthanized due to poor body conditions for humane reasons. (A) Percentage of CLL cases detected with age of Par-4+/+Tcl1 and Par-4−/−Tcl1 cohorts over time. P = .0002 comparing the 2 curves by log-rank test (n = 16, Par-4+/+Eμ-Tcl1; n = 10, Par-4−/−Tcl1). (B) Effect of Par-4 loss on the survival of Eµ-Tcl1 mice. Survival curve represents a total of 17 Par-4+/+ Tcl1 mice and 9 Par-4−/−Tcl1 mice. P = .0472 comparing the 2 curves by log-rank test. (C) Tissues from Par-4−/−Tcl1 mice were harvested and expression of Par-4 and Tcl1 proteins was determined by western blot analysis. Par-4 was detected in the spleen of Par-4+/+Tcl1 mouse, but was not present in any of the tissues of the Par-4−/−Tcl1 mouse. (D) Spleens from multiple Eμ-Tcl1 mice and Par-4−/−Tcl1 mice were harvested and total protein was isolated. Immunoblots were probed for Par-4 and p21. Protein expression was normalized to β-actin. LN, lymph node; Mesen. LN, mesenteric lymph node.
Primary human CLL cells express high levels of Par-4 regulated through BCR signaling
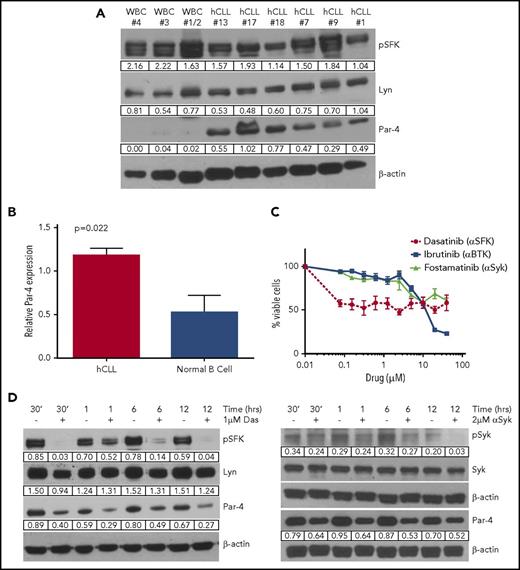
Characteristics of donor primary human CLL peripheral blood samples are summarized in Table 1. CLL status was confirmed through CD5+CD19+ staining by flow cytometry (supplemental Figure 13). Immunoblot analysis showed constitutive BCR signaling as reflected by p-SFK and total Lyn expression (Figure 7A). CLL cell samples expressed significantly higher levels of Par-4 protein when compared with normal donor cells. We also compared Par-4 expression in CLL cell samples to purified normal B cells and confirmed that malignant B cells express higher levels of Par-4 (Figure 7B). The Par-4 gene has been mapped to the middle portion of chromosome 12, and trisomy 12 is associated with ∼16% of CLL cases.39-41 There does not appear to be a gene dosage effect on Par-4 expression in trisomy 12 patients but more samples are needed to confirm this conclusion (supplemental Figure 14A). Par-4 expression was found to be slightly higher in the U-CLL patient samples compared with M-CLL but this difference was not statistically significant (supplemental Figure 14B). Survival of human CLL cells was also dependent on BCR signaling as treatment with inhibitors of SFK, Syk, and Btk kinases led to cell death (Figure 7C). Treatment of human CLL cells with dasatinib, fostamatinib, and ibrutinib led to a decrease in respective kinase activity and a downregulation of Par-4 protein expression (Figure 7D; supplemental Figure 15). Thus, human CLL cells have constitutive BCR signaling and inhibition of this signaling pathway regulates the expression of Par-4 that appears to promote CLL growth.
Human CLL patient information
| Patient no. . | Age, y . | Treated . | CD38+ . | Zap70+ . | IgVH mutation . | Cytogenetics . | WBC, × 109/L . |
|---|---|---|---|---|---|---|---|
| UK hCLL | |||||||
| 1 | 76 | No | Positive | U-CLL | Trisomy 12 | 19.8 | |
| 2 | 82 | No | Negative | Negative | M-CLL | 20.7 | |
| 3 | 56 | No | U-CLL | ||||
| 4 | 36 | No | Negative | M-CLL | 13q deletion | 38.3 | |
| 6 | 46 | No | Negative | Negative | M-CLL | 13q deletion | 41 |
| 7 | 52 | Yes | M-CLL | 15.2 | |||
| 8 | 69 | No | Positive | M-CLL | Normal | 29 | |
| 9 | 57 | Yes | U-CLL | 40 | |||
| 10 | 62 | Yes | M-CLL | 11.7% 17p deleted cells | 83.7 | ||
| 11 | 69 | No | M-CLL | Trisomy 12, 13q deletion | 30.2 | ||
| 13 | 70 | No | M-CLL | 17.4 | |||
| 14 | 63 | No | Positive | Positive | M-CLL | Trisomy 12 | 34.8 |
| 15 | 53 | Yes | Positive | Positive | U-CLL | 13q deletion | 12.1 |
| 16 | 55 | Yes | M-CLL | 6 | |||
| 17 | 76 | Yes | M-CLL | 36.6 | |||
| 18 | 80 | Yes | U-CLL | 142 | |||
| 20 | 57 | No | U-CLL | 7.4 | |||
| 21 | 78 | No | M-CLL | 45.9 | |||
| 22 | 74 | No | Negative | Negative | M-CLL | 11.3 | |
| 23 | 77 | Yes* | Negative | Negative | M-CLL | 11 | |
| 24 | 71 | No | Negative | Negative | M-CLL | 7.7 | |
| 25 | 70 | No | Negative | Negative | M-CLL | 23 | |
| 26 | 62 | No | Negative | Positive | M-CLL | 62.5 | |
| 27 | 64 | No | Negative | Positive | U-CLL | 9.4 | |
| 28 | 58 | Yes* | Negative | Positive | U-CLL | 8.4 | |
| 29 | 80 | Yes* | Positive | Positive | U-CLL | 39.6 | |
| 30 | 52 | No | Negative | Positive | U-CLL | 68.2 | |
| 31 | 66 | No | Negative | Negative | U-CLL | 20.4 | |
| OSU hCLL | |||||||
| 1 | 40 | M-CLL | 13q deletion | 88.2 | |||
| 2 | 59 | M-CLL | 13q deletion | 22.6 | |||
| 3 | M-CLL | 13q deletion | 99.1 | ||||
| 4 | 51 | M-CLL | Trisomy 12 | 148.8 | |||
| 5 | 71 | U-CLL | 138 | ||||
| 6 | U-CLL | 17p deletion; 13q deletion | 152 | ||||
| 7 | U-CLL | 13q deletion; 11q deletion | 196.6 | ||||
| 8 | 57 | U-CLL | 13q deletion; 2.5% trisomy 12 | 38.2 | |||
| 9 | M-CLL | 106.9 | |||||
| 10 | M-CLL | 13q deletion | 221.3 | ||||
| 11 | 61 | M-CLL | 78.2 | ||||
| 12 | 68 | M-CLL | 13q deletion | 170.9 | |||
| 13 | 72 | U-CLL | Trisomy 12 | 135.2 | |||
| 14 | 61 | U-CLL | 17p deletion; 13q deletion | 87 | |||
| 15 | 55 | U-CLL | 157.2 | ||||
| 16 | 67 | U-CLL | Trisomy 12 | 330.7 |
| Patient no. . | Age, y . | Treated . | CD38+ . | Zap70+ . | IgVH mutation . | Cytogenetics . | WBC, × 109/L . |
|---|---|---|---|---|---|---|---|
| UK hCLL | |||||||
| 1 | 76 | No | Positive | U-CLL | Trisomy 12 | 19.8 | |
| 2 | 82 | No | Negative | Negative | M-CLL | 20.7 | |
| 3 | 56 | No | U-CLL | ||||
| 4 | 36 | No | Negative | M-CLL | 13q deletion | 38.3 | |
| 6 | 46 | No | Negative | Negative | M-CLL | 13q deletion | 41 |
| 7 | 52 | Yes | M-CLL | 15.2 | |||
| 8 | 69 | No | Positive | M-CLL | Normal | 29 | |
| 9 | 57 | Yes | U-CLL | 40 | |||
| 10 | 62 | Yes | M-CLL | 11.7% 17p deleted cells | 83.7 | ||
| 11 | 69 | No | M-CLL | Trisomy 12, 13q deletion | 30.2 | ||
| 13 | 70 | No | M-CLL | 17.4 | |||
| 14 | 63 | No | Positive | Positive | M-CLL | Trisomy 12 | 34.8 |
| 15 | 53 | Yes | Positive | Positive | U-CLL | 13q deletion | 12.1 |
| 16 | 55 | Yes | M-CLL | 6 | |||
| 17 | 76 | Yes | M-CLL | 36.6 | |||
| 18 | 80 | Yes | U-CLL | 142 | |||
| 20 | 57 | No | U-CLL | 7.4 | |||
| 21 | 78 | No | M-CLL | 45.9 | |||
| 22 | 74 | No | Negative | Negative | M-CLL | 11.3 | |
| 23 | 77 | Yes* | Negative | Negative | M-CLL | 11 | |
| 24 | 71 | No | Negative | Negative | M-CLL | 7.7 | |
| 25 | 70 | No | Negative | Negative | M-CLL | 23 | |
| 26 | 62 | No | Negative | Positive | M-CLL | 62.5 | |
| 27 | 64 | No | Negative | Positive | U-CLL | 9.4 | |
| 28 | 58 | Yes* | Negative | Positive | U-CLL | 8.4 | |
| 29 | 80 | Yes* | Positive | Positive | U-CLL | 39.6 | |
| 30 | 52 | No | Negative | Positive | U-CLL | 68.2 | |
| 31 | 66 | No | Negative | Negative | U-CLL | 20.4 | |
| OSU hCLL | |||||||
| 1 | 40 | M-CLL | 13q deletion | 88.2 | |||
| 2 | 59 | M-CLL | 13q deletion | 22.6 | |||
| 3 | M-CLL | 13q deletion | 99.1 | ||||
| 4 | 51 | M-CLL | Trisomy 12 | 148.8 | |||
| 5 | 71 | U-CLL | 138 | ||||
| 6 | U-CLL | 17p deletion; 13q deletion | 152 | ||||
| 7 | U-CLL | 13q deletion; 11q deletion | 196.6 | ||||
| 8 | 57 | U-CLL | 13q deletion; 2.5% trisomy 12 | 38.2 | |||
| 9 | M-CLL | 106.9 | |||||
| 10 | M-CLL | 13q deletion | 221.3 | ||||
| 11 | 61 | M-CLL | 78.2 | ||||
| 12 | 68 | M-CLL | 13q deletion | 170.9 | |||
| 13 | 72 | U-CLL | Trisomy 12 | 135.2 | |||
| 14 | 61 | U-CLL | 17p deletion; 13q deletion | 87 | |||
| 15 | 55 | U-CLL | 157.2 | ||||
| 16 | 67 | U-CLL | Trisomy 12 | 330.7 |
Blank cells indicate that relevant information is not available. Total peripheral white blood cell (WBC) counts at the time of collection are indicated. Cytogenetics, CD38+, Zap70+ are indicated if known through previous diagnosis or by flow cytometry. CD38+ distinction was determined by >30% CD38+ cells of leukemic clone.23,58 Patient samples were collected at the University of Kentucky Markey Cancer Center (UK hCLL) or as indicated at The Ohio State (OSU hCLL) Brown Comprehensive Cancer Center.
Patient has received previous therapy but not on current treatment.
Primary human CLL cells have high levels of Par-4 protein expression. (A) Immunoblot analysis of activated SFK and Par-4 protein expression levels in primary human CLL (hCLL) samples compared with the whole peripheral blood lysate of normal donors (WBC). Protein values are normalized to β-actin. (B) Cell lysates of purified B cells from CLL patients (n = 4) and from healthy donors (n = 4) were probed for Par-4. The bar graph represents Par-4 protein values normalized to GAPDH expression. P = .022 determined by Student t test. (C) Survival curves of hCLL cells treated with BCR-signaling inhibitors. Assay performed in triplicate. Error bars represent SEM. (D) Primary human peripheral blood CLL cells (97.7% CD5+CD19+) were treated with 1 μM dasatinib (left) and 5 μM fostamatinib (right). p-SFK and Syk levels are normalized to their respective total protein levels. Total Syk, Lyn, and Par-4 protein expression levels are normalized to β-actin.
Primary human CLL cells have high levels of Par-4 protein expression. (A) Immunoblot analysis of activated SFK and Par-4 protein expression levels in primary human CLL (hCLL) samples compared with the whole peripheral blood lysate of normal donors (WBC). Protein values are normalized to β-actin. (B) Cell lysates of purified B cells from CLL patients (n = 4) and from healthy donors (n = 4) were probed for Par-4. The bar graph represents Par-4 protein values normalized to GAPDH expression. P = .022 determined by Student t test. (C) Survival curves of hCLL cells treated with BCR-signaling inhibitors. Assay performed in triplicate. Error bars represent SEM. (D) Primary human peripheral blood CLL cells (97.7% CD5+CD19+) were treated with 1 μM dasatinib (left) and 5 μM fostamatinib (right). p-SFK and Syk levels are normalized to their respective total protein levels. Total Syk, Lyn, and Par-4 protein expression levels are normalized to β-actin.
Discussion
Previously, Par-4 has been defined as a well-known tumor suppressor that is downregulated in several cancers and can selectively induce and/or sensitize cancer cells to apoptosis.42 This quality has made Par-4 an attractive potential therapeutic target in a variety of cancers, yet there are very few studies investigating its regulation. In this study, we made a surprising discovery that Par-4 expression in CLL is regulated by constitutive BCR signaling that promotes the growth of CLL.
Both mouse Eμ-Tcl1 and primary human CLL cells express high amounts of Par-4 compared with normal B-cell subsets and whole peripheral blood samples, respectively. Increased expression levels led to concerns regarding whether Par-4 expressed in CLL cells was functional but we confirmed that Par-4 was found in both the cytoplasm and the nucleus of CLL cells and was secreted functionally to induce apoptosis of other cancer cell lines. Previous studies have shown a direct interaction between p53 activation and elevated Par-4 secretion.43 p53 is mutated in the Mec-1 cell line, suggesting that Par-4 regulation is independent of p53 functional activity.44 Although the interplay between p53 and p21 expression is well documented,45 due to the mutation status of p53, we hypothesize that an alternative pathway may be activated with Par-4 loss and the subsequent p21 induction. Preliminary studies in our laboratory suggest that Par-4 is not mutated in Mec-1 cells. Knockdown studies targeting Par-4 in CLL generated a distinct phenotype that delayed the growth of CLL, which led us to investigate roles of Par-4 other than its tumor suppressor function.
Recently, cyclin-dependent kinase inhibitors are being investigated for the treatment of leukemia due to their success in solid tumors.46 Dinaciclib, a cyclin-dependent kinase inhibitor, reduced survival of CLL cells by inhibiting key oncogenic pathways.47 Our studies introduce Par-4 as a factor involved in the cell cycle through connection with p21 in CLL cells as reduced expression of Par-4 led to a halt in the G1/S transition of the cell cycle and a reciprocal induction of p21 expression. Interestingly, CLL cells in the proliferation centers located in the lymph node highly express cyclin D2, which is involved in the transition from the G0/G1 to the S phase of cell cycle, whereas p21 is downregulated in actively cycling cells.48,49 Studies investigating the interaction of Par-4 and cyclin D2 are warranted to elucidate the potential progrowth role of Par-4 in CLL. Another study recently investigated the relationship between Par-4 and p21 after inducing endoplasmic reticulum stress with the treatment of a natural product derivative, 3-Azido withaferin A (3-AWA).50 Induction of endoplasmic reticulum stress via 3-AWA treatment in prostate and colon cancer cells increased Par-4 expression and decreased p21. Results suggested that the loss of activated Akt allowed for increase in Par-4 expression, which then promoted apoptosis. The reduced expression of p21 after 3-AWA treatment was found to be mediated by proapoptotic Jun N-terminal kinase (JNK) activation. JNK is a key player in stress-induced cell death in both normal and solid tumor cancers.51 Conversely, our laboratory has discovered that JNK is required for survival and proliferation of malignant B-lymphoma cells, suggesting a dual role for this kinase in solid vs lymphoid cancers,52 which was shown to depend on the CARD11/Bcl10/Malt1 pathways with negative regulation by DUSP4.53,54 The diverse roles that JNK potentially plays in CLL vs epithelial cell tumors may be contributing to the progrowth function of Par-4 in CLL via p21 suppression.
To determine the effect of Par-4 on CLL growth in vivo, we used Par-4−/− mice, which were deficient for Par-4 in all tissues. Introduction of Par-4 deficiency delayed CLL development in the Eµ-Tcl1 mice and also prolonged their survival compared with Par-4+/+Tcl1 mice. Because these are whole-body knockouts for Par-4, the lack of Par-4 in the microenvironment and other cell types could also be playing a role in the delayed CLL growth observed. The Eμ-Tcl1 mice lacking Par-4 had an average lifespan of about 12 months, which was more than the Par-4+/+Tcl1 mice, although they developed CLL-like disease without indication of other tumors previously observed in the Par-4−/− animals.10 The characteristics of CLL in Par-4−/− mice were indistinguishable from Par-4–sufficient mice, demonstrating that Par-4 is not required for CLL but appears to promote disease growth, when present. Because CLL cells secrete Par-4, the CLL microenvironment could be affected by Par-4, which is currently being investigated. Additionally, we are generating B-cell–specific Par-4−/− mice to further demonstrate its B-cell–intrinsic role.
For the first time here, we show that constitutive Par-4 expression in CLL cells is regulated by BCR signaling by using small molecule inhibitors of upstream kinases activated by BCR as well as by shRNA-mediated knockdown of Lyn kinase (supplemental Figure 5) and siRNA targeting of CD79a (Figure 3B). When BCR signaling is engaged, Src family and Syk protein kinases are activated, which triggers activation of downstream signaling networks that include the Ras-MAPK (ERK) pathway and the PI3K pathway.33 Targeting ERK1/2 in Eμ-Tcl1 CLL cells resulted in a decrease in Par-4 expression, further confirming that Par-4 is regulated by survival signaling in malignant CLL cells. Thus, high levels of Par-4 coexisted with constitutive ERK activation in CLL cells, which is once again different from fibroblasts wherein Par-4 was shown to block ERK2 expression to prevent Ras transformation and Par-4 itself was inhibited by the Ras-MEK-ERK pathway.55 PC-3 prostate cancer cells also exhibit ERK1/2 activation despite expressing high levels of Par-4, which appears to be due to signaling upstream of ERK1/2 in PC-3 cells. PKCα and epidermal growth factor receptor signaling have both been found to be responsible for ERK1/2 signaling in PC-3 cells and could be masking the effect of ERK inhibition by Par-4.56 PKCα is found to be variably expressed in CLL cells and potentially could not compensate for the inhibition of ERK1/2 signaling on Par-4 expression,57 suggesting that Par-4 regulation by the Ras-ERK pathway may be tissue specific.
In summary, our studies demonstrate that constitutive Par-4 expression is regulated by the well-known tonic signaling via BCR in CLL cells, showing, for the first time, regulation of Par-4 expression by a cell surface receptor and that the constitutively expressed Par-4 is functional. Furthermore, our studies suggest that the tumor suppressor Par-4 may have a novel progrowth role in the context of CLL. Because Par-4 enhances the growth of CLL cells, it may be beneficial to CLL patients to inhibit Par-4. Current therapeutics for CLL target the BCR-signaling pathway, which may inadvertently reduce Par-4 expression. In future studies, it would be interesting to see whether combination therapy involving Par-4 inhibition would be more effective than inhibition of BCR signaling alone, especially for patients who develop resistance to such therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
A special thanks to Nikhil Hebbar for expertise and assistance in performing the Par-4 apoptosis assays. Another special thanks to the Markey Cancer Center Hematology-Oncology Clinic nurses for help in collecting patient samples.
This work was supported by the National Institutes of Health, National Cancer Institute (CA 165469) (N.M., V.M.R., and S.B.). M.K.M. and J.R.R. were supported by National Institutes of Health, National Cancer Institute T32 grant CA165990. This research was supported by the Biospecimen Procurement and Translational Pathology shared resource facilities of the University of Kentucky Markey Cancer Center (P30 CA177558). The Flow Cytometry & Cell Sorting core facility is supported in part by the Office of the Vice President for Research, the Markey Cancer Center, and a National Cancer Institute Center Core Support Grant (P30 CA177558) to the University of Kentucky Markey Cancer Center.
Authorship
Contribution: M.K.M. designed and performed experiments closely with the help of S.K.N. and S.S.A.; M.K.M. collected, analyzed, and interpreted data, performed statistical analysis, and wrote the manuscript; K.Z.O., S.K.N., K.L.P., J.P.C., J.R.R., J.T.G., and R.M. performed experiments, contributed to helpful discussions, and reviewed the manuscript; G.C.H. and R.A.F. contributed to obtaining human CLL samples and reviewed the manuscript; E.B.D. contributed by confirming patient information and reviewed the manuscript; C.W. provided additional help with statistical analysis and reviewed the manuscript; J.C.B. contributed to obtaining Eμ-Tcl1 mice and reviewed the manuscript; V.M.R. provided intellectual input and expertise on Par-4 and reviewed the manuscript; N.M. contributed to obtaining Eμ-Tcl1 mice, provided intellectual input, and reviewed the manuscript; and S.B. designed experiments, interpreted data, supervised the research, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Subbarao Bondada, University of Kentucky, 800 Rose St, 212 Combs Research Building, Lexington, KY 40536; e-mail: bondada@email.uky.edu.

![Figure 3. Inhibition and silencing of BCR signaling results in a decrease in Par-4 expression. (A) Mec-1 CLL cells were treated with control (Ctrl) or Lyn-specific shRNA lentivirus and selected through puromycin treatment. Cells were collected at day 17 and total protein and RNA was isolated. Left panel, Levels of Lyn protein. Right panel, Levels of Par-4 mRNA. (B) Human CLL cells were electroporated with CD79a siRNA and incubated for 24 hours. Total protein was isolated and probed for CD79a and Par-4. Western blot represents data from 1 CLL donor with average results for 4 other donors shown as histogram. Ctrl siRNA, n = 2; CD79a siRNA, n = 2. Error bars represent SEM. (C) Surface IgM levels were measured on human CLL cells electroporated with control and CD79a siRNA by flow cytometry (left, representative histogram for 1 patient; middle, average mean fluorescent intensity [MFI] of IgM expression for 4 donors). Total RNA was isolated from cells, and CD79a and Par-4 mRNA were measured by qRT-PCR. Expression values were normalized to human 18S expression performed in triplicate for 4 different donors. *P ≤ .05, **P ≤ .01, ***P ≤ .001 determined by the Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/26/10.1182_blood-2017-10-813931/4/m_blood813931f3.jpeg?Expires=1767802539&Signature=IMYlFWPjGCQ1nUOdm1EYpd~x06SkxiVLifwpCzKRSDfFqgXMWihTA1YerRjTS9Arl7cPant8003ehclLJhdc4M1APLWpvkPRTJ~dsXBsjcIB4vXBAyCCd1ax2UkdHjNs22XWkJDt9bus5sCHeR33hbQvsI0rbWeoNn-rMVPZD4zP5Rb~VyEIhpn6xBCSfkOqtJaTqhk2koK2SEaNuk6TrOL2QxULRpZ4xRs0aObXYqbJjHNUyMcwcntqH-mj7yNJstWNloiTzQ3Wkkrx7Fss2h~IGyFpnfScHnqBJ-47txrONaAFyH5TyBTwOC6YLNaoImKGaE1eUZqlzaXjfY9jdw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal