Key Points
CRISPR/Cas9-mediated disruption of a BCL11A binding site in HSCs of β-YAC mice results in the reactivation of γ-globin in erythrocytes.
Our approach for in vivo HSC genome editing that does not require HSC transplantation and myeloablation should simplify HSC gene therapy.
Abstract
Disorders involving β-globin gene mutations, primarily β-thalassemia and sickle cell disease, represent a major target for hematopoietic stem/progenitor cell (HSPC) gene therapy. This includes CRISPR/Cas9-mediated genome editing approaches in adult CD34+ cells aimed toward the reactivation of fetal γ-globin expression in red blood cells. Because models involving erythroid differentiation of CD34+ cells have limitations in assessing γ-globin reactivation, we focused on human β-globin locus-transgenic (β-YAC) mice. We used a helper-dependent human CD46-targeting adenovirus vector expressing CRISPR/Cas9 (HDAd-HBG-CRISPR) to disrupt a repressor binding region within the γ-globin promoter. We transduced HSPCs from β-YAC/human CD46–transgenic mice ex vivo and subsequently transplanted them into irradiated recipients. Furthermore, we used an in vivo HSPC transduction approach that involves HSPC mobilization and the intravenous injection of HDAd-HBG-CRISPR into β-YAC/CD46–transgenic mice. In both models, we demonstrated efficient target site disruption, resulting in a pronounced switch from human β- to γ-globin expression in red blood cells of adult mice that was maintained after secondary transplantation of HSPCs. In long-term follow-up studies, we did not detect hematological abnormalities, indicating that HBG promoter editing does not negatively affect hematopoiesis. This is the first study that shows successful in vivo HSPC genome editing by CRISPR/Cas9.
Introduction
The human β-globin locus is composed of 5 genes (ε, γ-A, γ-G, δ, and β). Expression of these genes is controlled by a single locus control region (LCR) in a developmental manner.1-3 In a process called globin switching, fetal hemoglobin expressed by γ-globin genes (HBG1 and HBG2) is nearly completely replaced by adult hemoglobin (β-globin) by ∼6 months postnatally. Disorders involving β-globin gene mutations (ie, sickle cell disease [SCD] and β-thalassemia) become symptomatic at this time. In hereditary persistence of fetal hemoglobin (HPFH), a benign genetic condition, mutations attenuate γ-to-β globin switching, causing high fetal globin (HbF) levels throughout life, thus alleviating the clinical manifestations of these disorders.4 This provided a rationale for gene therapy approaches of hemoglobinopathies aimed toward the reversal of globin switching (reviewed by Vinjamur et al5 and Psatha et al6 ). In this context, it has been shown that re-enacting HPFH mutations by creating large deletions within the β-globin locus7 or by introduction of mutations in the HBG promoters can increase the levels of HbF in erythroid cells.8-10 Of particular interest to us was a 13-bp deletion at position −114 to −102 with regard to the HBG1 and HBG2 transcription start sites. Traxler et al showed that lentivirus vector–mediated CRISPR/Cas9 gene transfer targeting the 13-bp region resulted in reactivation of γ-globin in cultures of an erythroid progenitor cell line (HUDEP-2) and in CD34+ cells after erythroid differentiation.9
However, the disadvantage of the CD34+/NSG xenotransplantation model is that it does not efficiently support human erythropoiesis, complicating the downstream analysis of globin gene editing in vivo.11 Moreover, in vitro erythroid differentiation of engrafted human hematopoietic stem/progenitor cell (HSPCs) in the presence of erythropoietin and other cytokines triggers the artificial activation of γ-globin expression in control settings.9,12 A more adequate model that allows for direct in vivo analysis of γ-globin reactivation are β-YAC mice; that is, mice carrying the human β-globin gene locus as a yeast artificial chromosome transgene.13 β-YAC mice have been used extensively in globin-switching studies, as well as for evaluation of several known HPFH mutations14-17 and γ-globin repressors.18,19
For CRISPR/Cas9 gene transfer, we used a nonintegrating helper-dependent adenovirus vector (HDAd5/35++) with high affinity to CD46, a receptor that is uniformly expressed on HSPCs and at higher levels than on more differentiated bone marrow and blood cells.20 We and other investigators have previously shown that CD46-targeting adenovirus vectors efficiently transduce primitive quiescent HSPCs from humans,20-23 nonhuman primates,24 and human CD46–transgenic mice.20 In contrast to recombinant adeno-associated virus and lentivirus vectors, HDAd5/35++ vector production does not require large-scale plasmid transfection, and a single HDAd5/35++ vector stock can be used for multiple manufacturing cycles.
Currently used ex vivo HSPC gene therapy is a complex and expensive procedure requiring extensive HSPC manipulation and transplantation expertise. Moreover, the intense myelo-conditioning/ablation required to reach clinically relevant HSPC engraftment levels increases toxicity and prolongs hospitalization. We developed a minimally invasive and readily translatable approach for in vivo HSPC gene delivery without leukapheresis, myeloablation, and HSPC transplantation.20,25 It involves injections of granulocyte colony-stimulating factor (G-CSF)/AMD3100 to mobilize HSPCs from the bone marrow into the peripheral blood stream and the intravenous injection of HDAd5/35++ vectors. We previously showed in adequate mouse models using an integrating HDAd5/35++ vector expressing GFP and mgmtP140K that HSPCs transduced in the periphery home back to the bone marrow where they persist long-term. Without a proliferative advantage, in vivo–transduced HSPCs do not efficiently exit the bone marrow and contribute to downstream differentiation. Short-term treatment of animals with O6BG/BCNU provides a proliferation stimulus to mgmtP140K gene–modified HSPCs and subsequent stable transgene expression in >80% of peripheral blood cells.26 Recently, we also demonstrated in vivo HSPC transduction with an HDAd5/35++-GFP vector in mobilized macaques.27
Here, we used a nonintegrating HDAd5/35++ vector for HSPC genome editing in vitro and in vivo with the goal to reactivate human γ-globin expression in red blood cells (RBCs) in β-YAC mice.
Materials and methods
HDAd5/35++ vectors
The construction of HDAd-HBG-CRISPR and HDAd-HBG-CRISPR/mgmt vectors is described in detail in supplemental Materials and methods, available on the Blood Web site.
CD34+ cell culture
CD34+ cells from G-CSF–mobilized adult donors were recovered from frozen stocks and incubated overnight in StemSpan H3000 (STEMCELL Technologies, Vancouver, BC, Canada) with penicillin/streptomycin, Flt3 ligand (25 ng/mL), interleukin-3 (10 ng/mL), thrombopoietin (2 ng/mL), and stem cell factor (25 ng/mL). Differentiation of human HSPCs into erythroid cells was done based on the protocol developed by Douay and Giarratana.28
Animal studies
All experiments involving animals were conducted in accordance with the institutional guidelines set forth by the University of Washington. Immunodeficient NSG mice (strain NOD/Shi-scid/IL-2Rγnull) and C57BL/6 mice were obtained from The Jackson Laboratory. C57BL/6-based transgenic mice that contained the human CD46 genomic locus and provide CD46 expression at a level and in a pattern similar to humans (human CD46+/+ mice) were described earlier.29 Transgenic mice carrying the wild-type 248-kb β-globin locus yeast artificial chromosome (β-YAC) were used.30 β-YAC mice were crossed with human CD46+/+ mice to obtain β-YAC+/−/CD46+/− mice for ex vivo studies and β-YAC+/−/CD46+/+ mice for in vivo HSPC-transduction studies.
Transplantation of human CD34+ cells into NSG mice.
NSG recipient mice received whole-body irradiation (300 rad). A total of 2.5 × 105 freshly collected whole bone marrow cells from nonirradiated NSG mice was mixed with 1 × 106 transduced human CD34+ cells and injected intravenously into recipient mice at 4 hours postirradiation. Animals were kept on Baytril for 4 weeks.
Bone marrow Lin− cell transplantation.
Recipients were 6-8-week-old female C57BL/6 mice. On the day of transplantation, recipient mice were irradiated (1000 rad). Four hours after irradiation, 1 × 106 Lin− cells were injected intravenously through the tail vein.
HSPC mobilization and in vivo transduction.
HSPCs were mobilized in mice by subcutaneous injections of human recombinant G-CSF (5 μg per mouse per day for 4 days) followed by a subcutaneous injection of AMD3100 (5 mg/kg) on day 5. In addition, animals received dexamethasone (10 mg/kg, intraperitoneally) 16 h and 2 h before virus injection. At 30 and 60 minutes after AMD3100 injection, animals were injected intravenously with HDAd-HBG-CRISPR through the retro-orbital plexus at a dose of 4 × 1010 viral particles (vp’s) per injection. Four weeks later, mice were injected with O6-BG (15 mg/kg, intraperitoneally) twice, 30 minutes apart. One hour after the second injection of O6-BG, mice were injected with BCNU (5 mg/kg, intraperitoneally). The BCNU dose was increased in the second cycle to 10 mg/kg.
Nucleofection, globin flow cytometry and high-performance liquid chromatography (HPLC), real-time reverse-transcription polymerase chain reaction (PCR) for globin messenger RNA (mRNA), mismatch sensitive nuclease assay T7E1 assay, deep sequencing of indels, magnetic activated cell sorting, progenitor colony assays, generation of HUDEP-2 clones, analysis of off-target sites, and statistical methods are described in supplemental Materials and methods.
Results
Target site cleavage and γ-globin reactivation after ex vivo transduction of human CD34+ cells with HDAd-HBG-CRISPR
We focused on a region associated with a known HPFH mutation, a 13-bp deletion at −102 to −114 (Figure 1A; supplemental Figure 1). Our single guide RNA (sgRNA) selection was based on the findings of Traxler et al, showing that targeting upstream rather than within the 13-bp deletion would have a greater effect on HbF reactivation.9 Based on 2 recent reports,31,32 which were not yet published at the time of our sgRNA design, this region overlaps with a binding site for the γ-globin repressor BCL11A (“TGACCA motif”) (Figure 1A). A corresponding HDAd-HBG-CRISPR vector (Figure 1B) was produced, and CRISPR/Cas9 cleavage activity and specificity were validated by T7E1 assay in peripheral blood CD34+ cells from healthy mobilized donors at day 3 after infection with HDAd-HBG-CRISPR. The HBG target site cleavage frequency in CD34+ cells from 2 donors was 23.8% and 20.8% (supplemental Figure 2). No cleavage activity was detected for the top 10 off-target sites in the human genomes (supplemental Figure 3A; supplemental Table 1). Erythroid differentiation was not negatively affected by HDAd-HBG-CRISPR transduction (supplemental Figure 4). The degree of enucleation and the expression of the erythroid differentiation markers CD71 and glycophorin A were comparable in in vitro–differentiated erythroid cells derived from untransduced and HDAd-HBG-CRISPR–transduced CD34+ cells.
Studies with human CD34+cells. (A) Schematic structure of globin locus with localization of the sgRNA in the promoters of the γ-globin genes, the CRISPR/Cas9 cleavage site (arrowhead), the 13-bp (−114 to −102) HPFH deletion (box), and the BCL11A-binding motif (“TGACCA” bold sequence). Note that only the distal TGACCA motif (red) within the indicated sequence is critical for γ-globin silencing.32 The CRISPR/Cas9 cleavage site is located 2 bp upstream of the BCL11A binding motif. ε, Gγ, Aγ, δ, and β are globin genes. (B) HDAd-HBG-CRISPR vector structure. The sgRNA gene is transcribed by PolIII from the U6 promoter, and the spCas9 gene is under the control of the EF1α promoter. Cas9 expression is controlled by miR-183-5p and miR-218-5p, which suppress Cas9 expression in HDAd producer cells but do not negatively affect Cas9 expression in CD34+ cells.35 The corresponding microRNA target sites (miR-T) were embedded into a 3′untranslated region (3′UTR). (C) Experimental design. CD34+ cells were transduced with HDAd-HBG-CRISPR at a multiplicity of infection of 2000 vp’s per cell. Two days later, cells were infected with HDAd–anti-CRISPR36 at the same multiplicity of infection to terminate CRISPR/Cas9 activity and, thus, minimize cytotoxicity to HSPCs. (The 2 anti-CRISPR peptides [AcrII4 and AcrII2] expressed from this vector are capable of binding to the CRISPR/Cas9 complex, thus blocking its activity.57 ) Sixteen hours after the last infection, CD34+ cells were transplanted into irradiated NSG mice. Untransduced cells were incubated for 2 days + 16 hours in the same medium as transduced cells. At week 10 after transplantation, human CD45+ cells were isolated from the bone marrow by magnetic-activated cell sorting and subjected to T7E1 assay and erythroid in vitro differentiation for globin analysis by flow cytometry and HPLC. (D) Target site cleavage frequency (measured by T7E1 assay) in human CD45+ cells isolated from bone marrow at week 10 after transplantation. Each lane is an individual mouse. (E) γ-Globin flow cytometry of cells after erythroid in vitro differentiation. (F) Ratio of γ-globin to adult α- or β-globin measured by HPLC after erythroid differentiation. n = 3. HS, DNase I hypersensitivity sites; untr, mice transplanted with untransduced CD34+ cells.
Studies with human CD34+cells. (A) Schematic structure of globin locus with localization of the sgRNA in the promoters of the γ-globin genes, the CRISPR/Cas9 cleavage site (arrowhead), the 13-bp (−114 to −102) HPFH deletion (box), and the BCL11A-binding motif (“TGACCA” bold sequence). Note that only the distal TGACCA motif (red) within the indicated sequence is critical for γ-globin silencing.32 The CRISPR/Cas9 cleavage site is located 2 bp upstream of the BCL11A binding motif. ε, Gγ, Aγ, δ, and β are globin genes. (B) HDAd-HBG-CRISPR vector structure. The sgRNA gene is transcribed by PolIII from the U6 promoter, and the spCas9 gene is under the control of the EF1α promoter. Cas9 expression is controlled by miR-183-5p and miR-218-5p, which suppress Cas9 expression in HDAd producer cells but do not negatively affect Cas9 expression in CD34+ cells.35 The corresponding microRNA target sites (miR-T) were embedded into a 3′untranslated region (3′UTR). (C) Experimental design. CD34+ cells were transduced with HDAd-HBG-CRISPR at a multiplicity of infection of 2000 vp’s per cell. Two days later, cells were infected with HDAd–anti-CRISPR36 at the same multiplicity of infection to terminate CRISPR/Cas9 activity and, thus, minimize cytotoxicity to HSPCs. (The 2 anti-CRISPR peptides [AcrII4 and AcrII2] expressed from this vector are capable of binding to the CRISPR/Cas9 complex, thus blocking its activity.57 ) Sixteen hours after the last infection, CD34+ cells were transplanted into irradiated NSG mice. Untransduced cells were incubated for 2 days + 16 hours in the same medium as transduced cells. At week 10 after transplantation, human CD45+ cells were isolated from the bone marrow by magnetic-activated cell sorting and subjected to T7E1 assay and erythroid in vitro differentiation for globin analysis by flow cytometry and HPLC. (D) Target site cleavage frequency (measured by T7E1 assay) in human CD45+ cells isolated from bone marrow at week 10 after transplantation. Each lane is an individual mouse. (E) γ-Globin flow cytometry of cells after erythroid in vitro differentiation. (F) Ratio of γ-globin to adult α- or β-globin measured by HPLC after erythroid differentiation. n = 3. HS, DNase I hypersensitivity sites; untr, mice transplanted with untransduced CD34+ cells.
For in vivo studies, CD34+ cells were transduced with HDAd-HBG-CRISPR and transplanted into irradiated NSG mice (Figure 1C). Engraftment, measured at week 10 after transplantation and based on human CD45+ cells in bone marrow mononuclear cells, was 10-20% in individual mice (supplemental Figure 2), which is in agreement with previous studies using GFP-expressing HDAd5/35++ vectors.33-37 At week 10 posttransplantation, the frequency of target site cleavage in human CD45+ cells isolated from bone marrow ranged from 19% to 25% in individual mice (Figure 1D) and was in the range of the pretransplant population (supplemental Figure 2). Next-generation sequencing of the HBG region in CD45+ cells showed that the majority of indels were 1-nt deletions or insertions within the 13-bp HPFH region (supplemental Figure 6). Upon erythroid in vitro differentiation of bone marrow–derived human CD45+ cells, the frequency of γ-globin+ cells was ∼50% in HDAd-HBG-CRISPR settings compared with ∼27% in erythroid cells from mice that were transplanted with untransduced CD34+ cells (Figure 1E). Furthermore, HPLC analyses showed that the ratio of γ-globin to adult α- and β-globin protein was increased compared with untransduced cells, indicating a partial switch from adult globin to fetal γ-globin, confirming that the HDAd-HBG-CRISPR vector is functional (Figure 1F).
Target site cleavage and γ-globin reactivation after ex vivo transduction of β-YAC/CD46 Lin− cells with HDAd-HBG-CRISPR
A major drawback of the CD34+/NSG model is that in vivo evaluation of γ-globin reactivation in mature circulating erythrocytes is impossible. Therefore, we used β-YAC mice that contain 248 kb of human DNA, including the complete 82-kb β-globin locus. To allow for HDAd5/35++ transduction, we crossed them with human CD46-transgenic mice that express the HDAd5/35++ receptor CD46 in a pattern similar to humans.29 Peripheral RBCs of adult β-YAC/CD46 mice expressed human β-globin, but only ∼1% were positive for γ-globin (supplemental Figure 7), recapitulating the human pattern of globin expression in RBCs. Infection of bone marrow Lin− cells from β-YAC/CD46 mice with the HDAd-HBG-CRISPR vector demonstrated genomic target site cleavage by T7E1 assay and indel sequencing (supplemental Figure 8). No cleavage activity was detected for the top 15 off-target sites in the mouse genomes (supplemental Figure 3B; supplemental Table 2). To study γ-globin reactivation, we transduced bone marrow Lin− cells from β-YAC/CD46 mice with the HDAd-HBG-CRISPR vector and transplanted them into lethally irradiated C57BL/6 mice (Figure 2A). Engraftment measured at week 10 in primary recipients (based on human CD46 expression in peripheral blood mononuclear cells [PBMCs]) was >95% in bone marrow (9 of 9 mice) and spleen and blood (8 of 9 mice), indicating that HDAd-HBG-CRISPR transduction did not negatively affect the viability of repopulating HSPCs (Figure 2B). Target site cleavage/repair in total bone marrow cells harvested at week 10 was analyzed by T7E1 assay (Figure 2C; supplemental Figure 9). The mean cleavage frequency was 33%, 22%, and 30% in blood, spleen, and bone marrow, respectively (Figure 2C, first 3 columns). It was similar in bone marrow Ter119+ (erythroid) and Ter119− cells. In progenitor colonies derived Lin− cells, the cleavage frequency was ∼23%. This suggests that genome editing occurred in multipotent progenitors and/or primitive hematopoietic stem cells (HSCs). The mean total indel frequency in the mice analyzed was 80% (ie, higher than the cleavage frequency measured by the T7E1 assay) (Figure 2D). The latter observation was also reported recently by other investigators.38 A total of 70% of indels was deletions ranging from 1 to 20 bp, and only 10% were 1-2-bp microinsertions (Figure 2E; supplemental Figure 10). All indels (100%) were within the −114 to −102 HPFH region, and 50% of all deletions disrupted/removed the TGACCA BCL11A binding motif. A complete list of indels in all mice is shown in supplemental Table 1.
Ex vivo transduction of β-YAC/CD46 Lin−cells with HDAd-HBG-CRISPR and subsequent transplantation. (A) Schematic diagram of the experiment. Bone marrow was harvested from β-YAC/CD46 mice, and Lin− cells were isolated by magnetic-activated cell sorting. Lin− cells were transduced with HDAd-HBG-CRISPR vector at a multiplicity of infection of 500 vp’s per cell (blue squares) or were left untransduced (empty red squares). After 1 day in culture, 1 × 106 transduced cells per mouse were transplanted into lethally irradiated C57BL/6 mice. Animals were euthanized at week 10, and bone marrow Lin− cells were transplanted into secondary recipients that were subsequently followed for 16 weeks. (B) Engraftment at week 10 based on the percentage of human CD46+ cells in mononuclear cells of blood, spleen, and bone marrow. (C) Percentage of HBG target site cleavage measured by T7E1 assay at week 10 after transplantation in the indicated samples. Each symbol represents an individual mouse. Cells from colonies were pooled, and genomic DNA was isolated. (The corresponding polyacrylamide gels for the graphs are shown in supplemental Figure 4.) (D) Percentage of total HBG indels obtained by deep sequencing of DNA from total bone marrow mononuclear cells at week 10 after transplantation. Each symbol is an individual animal. (E) Top 30 most frequent indels found in mouse #976 (indel percentage = 58%). The blue sequence shows the target of the sgRNA with the TGACCA BCL11A binding motif underlined. The horizontal bold black line indicates the −114 to −102 HPFH. The CRISPR/Cas9 cleavage site is marked by a red arrowhead. The right panels show the number and percentage of reads for the corresponding indel. A complete list of the indels in all 3 mice is provided in supplemental Table 3. CFU, colony-forming units; MNC, mononuclear cells.
Ex vivo transduction of β-YAC/CD46 Lin−cells with HDAd-HBG-CRISPR and subsequent transplantation. (A) Schematic diagram of the experiment. Bone marrow was harvested from β-YAC/CD46 mice, and Lin− cells were isolated by magnetic-activated cell sorting. Lin− cells were transduced with HDAd-HBG-CRISPR vector at a multiplicity of infection of 500 vp’s per cell (blue squares) or were left untransduced (empty red squares). After 1 day in culture, 1 × 106 transduced cells per mouse were transplanted into lethally irradiated C57BL/6 mice. Animals were euthanized at week 10, and bone marrow Lin− cells were transplanted into secondary recipients that were subsequently followed for 16 weeks. (B) Engraftment at week 10 based on the percentage of human CD46+ cells in mononuclear cells of blood, spleen, and bone marrow. (C) Percentage of HBG target site cleavage measured by T7E1 assay at week 10 after transplantation in the indicated samples. Each symbol represents an individual mouse. Cells from colonies were pooled, and genomic DNA was isolated. (The corresponding polyacrylamide gels for the graphs are shown in supplemental Figure 4.) (D) Percentage of total HBG indels obtained by deep sequencing of DNA from total bone marrow mononuclear cells at week 10 after transplantation. Each symbol is an individual animal. (E) Top 30 most frequent indels found in mouse #976 (indel percentage = 58%). The blue sequence shows the target of the sgRNA with the TGACCA BCL11A binding motif underlined. The horizontal bold black line indicates the −114 to −102 HPFH. The CRISPR/Cas9 cleavage site is marked by a red arrowhead. The right panels show the number and percentage of reads for the corresponding indel. A complete list of the indels in all 3 mice is provided in supplemental Table 3. CFU, colony-forming units; MNC, mononuclear cells.
The effect of HDAd-HBG-CRISPR–mediated genome editing (ie, the reactivation of human γ-globin expression) was measured in peripheral RBCs in mice at weeks 4, 6, and 8 posttransplantation (Figure 3A). On average, 15% of RBCs expressed human γ-globin as early as 4 weeks posttransplantation. The percentage of γ-globin+ RBCs increased significantly by week 8 (22%). γ-Globin expression was localized exclusively to erythroid (Ter-119+) cells in blood and bone marrow (Figure 3B-C). The frequency of γ-globin+ cells in Ter-119+ erythroid cells in the bone marrow was lower than in peripheral RBCs, probably because only <50% of bone marrow Ter-119+ cells were terminally differentiated (supplemental Figure 11), and because of the overall low globin content in nucleated erythroid progenitors.
Analysis of human γ-globin reactivation and hematological parameters after ex vivo transduction of β-YAC/CD46 Lin−cells with HDAd-HBG-CRISPR and subsequent transplantation. (A) Percentage of human γ-globin+ cells in peripheral blood RBCs measured by flow cytometry in blood samples taken at weeks 4, 6, and 8 after transplantation with untransduced Lin− cells (□) and HDAd-HBG-CRISPR–transduced Lin− cells (▪). *P < .05. (B-E) Analysis of samples collected at week 10 posttransplantation. (B) Percentage of human γ-globin+ cells in erythroid (Ter119+) and nonerythroid (Ter119−) cells in blood and bone marrow. (C) Representative flow cytometry samples for data shown in (B). (D) Relative human β-globin (HBB) and γ-globin (HBG) mRNA levels in mice transplanted with HDAd-HBG-CRISPR–transduced Lin− cells compared with mRNA levels in untransduced settings (taken as 1.0). (E) HPLC data. Percentage of human γ-globin protein relative to human β-globin protein. (F-H) Safety of ex vivo HDAd-HBG-CRISPR genome editing in mouse HSPCs. (F) Hematological parameters at week 10 include RBCs (M/μL), hemoglobin (hB; g/dL), mean corpuscular hemoglobin concentration (MCHC; g/dL), white blood cells (WBC; K/μL); neutrophils (NE; K/μL); lymphocytes (LY; K/μl); monocytes (MO; K/μl); eosinophils (EO; K/μL), and basophils (BA; K/μL), and platelets (K/μL). n = 3. (G) Cell composition in blood, spleen, and bone marrow at week 10 after transplantation. Shown is the percentage of lineage marker-positive cells (CD3+, CD19+, Gr-1+, Ter119+ cells) and HSPCs (LSK cells). (H) Colony-forming potential of bone marrow Lin− cells harvested at week 10 posttransplantation. Number of colonies that formed after plating of 2500 Lin− cells (left panel) and total number of cells pooled from colonies (right panel). Each point represents an individual animal. n.s., not significant.
Analysis of human γ-globin reactivation and hematological parameters after ex vivo transduction of β-YAC/CD46 Lin−cells with HDAd-HBG-CRISPR and subsequent transplantation. (A) Percentage of human γ-globin+ cells in peripheral blood RBCs measured by flow cytometry in blood samples taken at weeks 4, 6, and 8 after transplantation with untransduced Lin− cells (□) and HDAd-HBG-CRISPR–transduced Lin− cells (▪). *P < .05. (B-E) Analysis of samples collected at week 10 posttransplantation. (B) Percentage of human γ-globin+ cells in erythroid (Ter119+) and nonerythroid (Ter119−) cells in blood and bone marrow. (C) Representative flow cytometry samples for data shown in (B). (D) Relative human β-globin (HBB) and γ-globin (HBG) mRNA levels in mice transplanted with HDAd-HBG-CRISPR–transduced Lin− cells compared with mRNA levels in untransduced settings (taken as 1.0). (E) HPLC data. Percentage of human γ-globin protein relative to human β-globin protein. (F-H) Safety of ex vivo HDAd-HBG-CRISPR genome editing in mouse HSPCs. (F) Hematological parameters at week 10 include RBCs (M/μL), hemoglobin (hB; g/dL), mean corpuscular hemoglobin concentration (MCHC; g/dL), white blood cells (WBC; K/μL); neutrophils (NE; K/μL); lymphocytes (LY; K/μl); monocytes (MO; K/μl); eosinophils (EO; K/μL), and basophils (BA; K/μL), and platelets (K/μL). n = 3. (G) Cell composition in blood, spleen, and bone marrow at week 10 after transplantation. Shown is the percentage of lineage marker-positive cells (CD3+, CD19+, Gr-1+, Ter119+ cells) and HSPCs (LSK cells). (H) Colony-forming potential of bone marrow Lin− cells harvested at week 10 posttransplantation. Number of colonies that formed after plating of 2500 Lin− cells (left panel) and total number of cells pooled from colonies (right panel). Each point represents an individual animal. n.s., not significant.
Analysis of human γ-globin (HBG) and β-globin (HBB) mRNA levels in RBCs showed an approximately fivefold decrease in HBB mRNA and an ∼30-fold increase in HBG mRNA compared with animals that received untransduced Lin− cells from β-YAC/CD46 mice (Figure 3D). This indicates a reverse switch from adult (HBB) to fetal (HBG) globin.
The adult-to-fetal globin change was also observed at the protein level in RBCs (Figure 3E). The level of HBG chains was ∼35% of HBB chains in mice transplanted with genome-edited HSPCs compared with ∼2% in mice transplanted with untransduced cells.
Blood cell counts and hemoglobin content (Figure 3F), as well as lineage composition in the bone marrow (Figure 3G), were similar in mice that received untransduced and HDAd-HBG-CRISPR–transduced Lin− cells. In the bone marrow, the levels of Lin−/Sca1+/cKit+ (LSK) HSPCs (Figure 3G) and progenitor colony-forming cells (Figure 3H) were also comparable in both groups.
To further confirm that our approach resulted in genetic modification of primitive (long-term repopulating) HSCs, Lin− bone marrow cells from primary recipients collected at week 10 after transplantation were used for secondary transplantation into lethally irradiated C57BL/6 mice, which were then followed for 16 weeks. Engraftment was >90% in most mice and stable (supplemental Figure 12A-B). Cellular bone marrow composition was similar in control and HBG-promoter edited groups (supplemental Figure 12C). The frequency of γ-globin RBCs in peripheral blood ranged from 7% to 30% and was stable (supplemental Figure 12D). At week 16, erythroid-restricted γ-globin expression was maintained (supplemental Figure 12E). The HBG/HBB ratio was similar to what we observed in the primary transplant recipients (supplemental Figure 12F).
In summary, these data suggest that our vector targets long-term repopulating cells and that HBG promoter modification results in stable reactivation of γ-globin in RBCs without hematological side effects.
In vivo HDAd-HBG-CRISPR HSPC transduction of mobilized β-YAC/CD46 cells
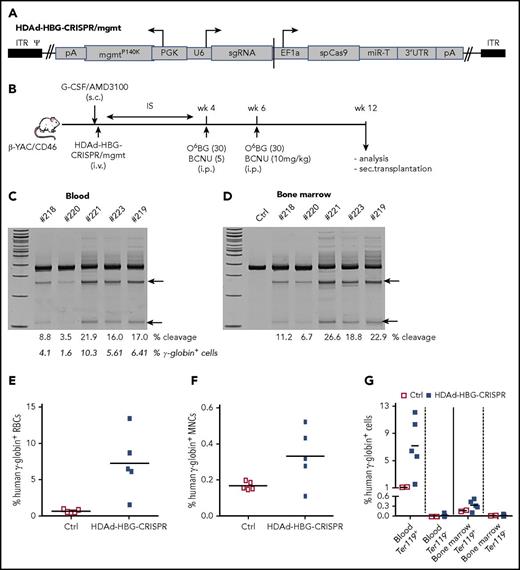
We next tested in vivo HSPC genome editing after intravenous injection into mobilized mice. In the case of a nonintegrating episomal vector, such as HDAd-HBG-CRISPR, cell division will lead to an abortive loss of vector genomes and, consequently, CRISPR/Cas9 expression, which is a desirable scenario, because genome editing requires only short-term CRISPR/Cas9 activity.39 Therefore, we incorporated a mgmtP140K expression cassette into the HDAd-HBG-CRISPR vector (Figure 4A). For in vivo HSPC transduction, HDAd-HBG-CRISPR/mgmt was intravenously injected into G-CSF/AMD3100–mobilized β-YAC/CD46 mice (Figure 4B). At week 4 after mobilization, when the cellular composition in the bone marrow returned to premobilization parameters,20 mice were treated with 2 cycles of O6BG/BCNU 2 weeks apart and analyzed 6 weeks later (week 12 post–in vivo transduction). Target site–specific genome cleavage measured by T7E1 assay ranged from 3.5% to 21.9% (Figure 4C) and from 6.7% to 26.6% (Figure 4D) in total blood and bone marrow cells, respectively. As expected from studies with integrating HDAd-GFP vectors,26 in vivo HDAd-HBG-CRISPR transduction of β-YAC/CD46 mice without O6BG/BCNU treatment resulted in target site–specific cleavage signals in the bone marrow but not in PBMCs (supplemental Figure 13). In β-YAC/CD46 mice that received in vivo transduction and O6BG/BCNU treatment, target site cleavage in HSPCs resulted in reactivation of γ-globin expression in up to 13% of RBCs (Figure 4E), and the percentage of γ-globin+ RBCs correlated with the genomic site cleavage frequency (Figure 4C, second row of numbers). As seen in the ex vivo study, the mean percentage of γ-globin+ cells in total bone marrow cells was 0.35%. γ-Globin expression was specific to erythroid Ter119+ blood and bone marrow cells (Figure 4G; supplemental Figure 14).
In vivo HDAd-HBG-CRISPR/mgmt transduction of mobilized HSPCs in β-YAC/CD46 mice. (A) Structure of HDAd-HBG-CRISPR/mgmt vector. In addition to the HBG CRISPR/Cas9 cassette, the vector contained a PGK promoter-driven mgmtP140K gene. (B) Schematic diagram of the experiment. β-YAC/CD46 mice were mobilized by subcutaneous injections of G-CSF and AMD3100 and were subsequently injected intravenously with HDAd-HBG/CRISPR/mgmt. To avoid immune responses against the bacterial Cas9 and human mgmtP140K proteins expressed from the episomal vector, mice received immunosuppressive drugs (IS) for 4 weeks. At weeks 4 and 6, mice were injected intraperitoneally with O6BG/BCNU at the indicated doses. Animals were euthanized at week 12 after in vivo transduction, and tissues were analyzed. Bone marrow Lin− cells were transplanted into C57BL/6 mice. T7E1 assay on total blood (C) and bone marrow (D) mononuclear cells at week 12 posttransduction. Specific cleavage products are indicated by arrows. The numbers above the gels are ID tags of individual mice. The numbers below the gels are the percentages of target site cleavage. The percentage of γ-globin+ cells in RBCs is also indicated. (E) Percentage of human γ-globin+ cells in peripheral blood RBCs measured by flow cytometry in blood samples. (F) Percentage of human γ-globin+ cells in total bone marrow mononuclear cells. (G) Percentage of human γ-globin+ cells in erythroid (Ter119+) and nonerythroid (Ter119−) cells in blood and bone marrow. Ctrl, mock-transduced mice.
In vivo HDAd-HBG-CRISPR/mgmt transduction of mobilized HSPCs in β-YAC/CD46 mice. (A) Structure of HDAd-HBG-CRISPR/mgmt vector. In addition to the HBG CRISPR/Cas9 cassette, the vector contained a PGK promoter-driven mgmtP140K gene. (B) Schematic diagram of the experiment. β-YAC/CD46 mice were mobilized by subcutaneous injections of G-CSF and AMD3100 and were subsequently injected intravenously with HDAd-HBG/CRISPR/mgmt. To avoid immune responses against the bacterial Cas9 and human mgmtP140K proteins expressed from the episomal vector, mice received immunosuppressive drugs (IS) for 4 weeks. At weeks 4 and 6, mice were injected intraperitoneally with O6BG/BCNU at the indicated doses. Animals were euthanized at week 12 after in vivo transduction, and tissues were analyzed. Bone marrow Lin− cells were transplanted into C57BL/6 mice. T7E1 assay on total blood (C) and bone marrow (D) mononuclear cells at week 12 posttransduction. Specific cleavage products are indicated by arrows. The numbers above the gels are ID tags of individual mice. The numbers below the gels are the percentages of target site cleavage. The percentage of γ-globin+ cells in RBCs is also indicated. (E) Percentage of human γ-globin+ cells in peripheral blood RBCs measured by flow cytometry in blood samples. (F) Percentage of human γ-globin+ cells in total bone marrow mononuclear cells. (G) Percentage of human γ-globin+ cells in erythroid (Ter119+) and nonerythroid (Ter119−) cells in blood and bone marrow. Ctrl, mock-transduced mice.
Analysis of mRNA levels in RBCs of in vivo mock-transduced and HDAd-HBG-CRISPR– transduced mice showed that HBG-promoter editing decreased HBB mRNA levels approximately threefold and increased HBG levels ∼10-fold (Figure 5A), resulting in an ∼12-fold increase in the ratio of HBG/HBB mRNA (Figure 5B). This is also reflected at the level of HBG and HBB protein chains (Figure 5C). Using a modified HPLC program that allowed for a better resolution of Gγ and Aγ chains (supplemental Figure 15), we showed that untransduced β-YAC/CD46 mice expressed low-level Gγ but no Aγ. In vivo transduction with HDAd-HBG-CRISPR led to an increase in Gγ and the appearance of Aγ. The relative increase (compared with human β-globin) of Aγ was greater than that of Gγ (Figure 5C).
β-Globin to γ-globin switch in in vivo–transduced β-YAC/CD46 mice and hematological parameters. (A) Relative human β-globin (HBB) and γ-globin (HBG) mRNA levels in peripheral blood RBCs and bone marrow erythroid Ter-119+ cells. mRNA levels in untransduced mice were taken as 1.0. (B) Percentage of HBG mRNA of human HBB mRNA (untransduced, empty red squares; HDAd-HBG-CRISPR–transduced, blue squares). (C) HPLC data. Percentage of human Gγ- and Aγ-globin protein relative to human β-globin protein in RBCs from untransduced and HDAd-HBG-CRISPR mice (week 12 after transduction). (D-E). Hematological safety of in vivo HDAd-HBG-CRISPR genome editing. (D) Cellular composition in blood (CD3+, CD19+, Gr-1+), spleen (CD3+, CD19+, Gr-1+), and bone marrow (CD3+, CD19+, Gr-1+, Ter119+, LSK) at week 12 after in vivo transduction. Shown is the percentage of lineage marker–positive cells (CD3+, CD19+, Gr-1+, Ter119+ cells) and HSPCs (LSK cells). (E) Colony-forming potential of bone marrow Lin− cells harvested at week 12 after in vivo HSPC transduction of β-YAC/CD46 mice. Number of colonies that formed after plating of 2500 Lin− cells (left panel) and total number of cells pooled from colonies (right panel). Each point is an individual animal.
β-Globin to γ-globin switch in in vivo–transduced β-YAC/CD46 mice and hematological parameters. (A) Relative human β-globin (HBB) and γ-globin (HBG) mRNA levels in peripheral blood RBCs and bone marrow erythroid Ter-119+ cells. mRNA levels in untransduced mice were taken as 1.0. (B) Percentage of HBG mRNA of human HBB mRNA (untransduced, empty red squares; HDAd-HBG-CRISPR–transduced, blue squares). (C) HPLC data. Percentage of human Gγ- and Aγ-globin protein relative to human β-globin protein in RBCs from untransduced and HDAd-HBG-CRISPR mice (week 12 after transduction). (D-E). Hematological safety of in vivo HDAd-HBG-CRISPR genome editing. (D) Cellular composition in blood (CD3+, CD19+, Gr-1+), spleen (CD3+, CD19+, Gr-1+), and bone marrow (CD3+, CD19+, Gr-1+, Ter119+, LSK) at week 12 after in vivo transduction. Shown is the percentage of lineage marker–positive cells (CD3+, CD19+, Gr-1+, Ter119+ cells) and HSPCs (LSK cells). (E) Colony-forming potential of bone marrow Lin− cells harvested at week 12 after in vivo HSPC transduction of β-YAC/CD46 mice. Number of colonies that formed after plating of 2500 Lin− cells (left panel) and total number of cells pooled from colonies (right panel). Each point is an individual animal.
Analyses performed at week 12 (6 weeks after the last O6BG/BCNU injection) demonstrated that in vivo HSPC transduction of mobilized β-YAC/CD46 mice with HDAd-HBG-CRISPR and subsequent treatment with O6BG/BCNU do not change hematological parameters (data not shown), the cellular composition of bone marrow fractions (Figure 5D), or the colony-forming capacity of bone marrow Lin− cells (Figure 5E) compared with untransduced mice. Notably, we recently reported transient leukopenia/thrombocytopenia, associated with O6BG/BCNU injection, that resolved by day 14 after the last treatment.26 Overall, our data indicate that the in vivo transduction/selection approach does not negatively affect in vivo hematopoiesis in the long-term.
To further assess the safety and efficacy of transduction of long-term repopulating HSPCs using the in vivo approach, studies were performed in secondary recipients. Bone marrow Lin− cells harvested from β-YAC/CD46 mice at week 12 after in vivo transduction and from untransduced mice were transplanted into lethally irradiated C57BL/6 mice, which were then followed for 16 weeks. Notably, secondary recipients did not receive O6BG/BCNU treatment or immunosuppression. Engraftment rates of transplanted cells in blood, spleen, and bone marrow were ∼100% for transduced and untransduced settings (Figure 6A-B). The percentage of γ-globin+ peripheral RBCs reached an average of ∼10% at week 6 posttransplantation and was stable until week 16 (Figure 6C). The percentage of γ-globin+ RBCs and the target site cleavage efficacy (Figure 6D) were significantly higher (P < .05) than in primary transduced mice. γ-Globin expression was restricted to erythroid Ter119+ cells (Figure 6E). The levels of γ-globin in RBCs measured at the mRNA level (Figure 6F) and protein level (Figure 6G) were comparable to those seen in primary mice. No HDAd-HBG-CRISPR–related changes in bone marrow composition were observed in secondary recipients at week 16 after transplantation (Figure 6H).
Analysis of secondary recipients (in vivo HSPC transduction approach). Bone marrow Lin− cells harvested from in vivo–transduced β˗YAC/CD46 mice at week 12 after transduction were transplanted into lethally irradiated C57BL/6 mice. Secondary recipients were followed for 16 weeks. (A) Engraftment measured in blood samples at the indicated time points based on the percentage of human CD46+ cells in PBMCs. (B) Engraftment at week 16 based on the percentage of human CD46+ cells in mononuclear cells of blood, spleen, and bone marrow (BM). (C) Human γ-globin expression in secondary recipients. Shown is human γ-globin expression on RBCs from week 4 to week 16 after transplantation. (D) T7E1 assay on total blood and bone marrow mononuclear cells at week 16 posttransplantation. Specific cleavage products are indicated by arrows. The numbers above the gels are ID tags of individual mice. The numbers below the gels are the percentages of target site cleavage. For blood cells, the percentage of γ-globin+ cells in RBCs is also indicated. (E) Percentage of human γ-globin+ cells in erythroid (Ter119+) and nonerythroid (Ter119−) cells in blood and bone marrow (week 16 posttransplantation). (F) Percentage of HBG mRNA of human HBB mRNA. (G) HPLC data. Percentage of human Gγ- and Aγ-globin protein relative to human β-globin protein in RBCs from untransduced and HDAd-HBG-CRISPR mice (week 16 posttransplantation). (H) Cellular composition in blood (CD3+, CD19+, Gr-1+), spleen (CD3+, CD19+, Gr-1+), and bone marrow (CD3+, CD19+, Gr-1+, Ter119+, LSK) at week 16 after transplantation.
Analysis of secondary recipients (in vivo HSPC transduction approach). Bone marrow Lin− cells harvested from in vivo–transduced β˗YAC/CD46 mice at week 12 after transduction were transplanted into lethally irradiated C57BL/6 mice. Secondary recipients were followed for 16 weeks. (A) Engraftment measured in blood samples at the indicated time points based on the percentage of human CD46+ cells in PBMCs. (B) Engraftment at week 16 based on the percentage of human CD46+ cells in mononuclear cells of blood, spleen, and bone marrow (BM). (C) Human γ-globin expression in secondary recipients. Shown is human γ-globin expression on RBCs from week 4 to week 16 after transplantation. (D) T7E1 assay on total blood and bone marrow mononuclear cells at week 16 posttransplantation. Specific cleavage products are indicated by arrows. The numbers above the gels are ID tags of individual mice. The numbers below the gels are the percentages of target site cleavage. For blood cells, the percentage of γ-globin+ cells in RBCs is also indicated. (E) Percentage of human γ-globin+ cells in erythroid (Ter119+) and nonerythroid (Ter119−) cells in blood and bone marrow (week 16 posttransplantation). (F) Percentage of HBG mRNA of human HBB mRNA. (G) HPLC data. Percentage of human Gγ- and Aγ-globin protein relative to human β-globin protein in RBCs from untransduced and HDAd-HBG-CRISPR mice (week 16 posttransplantation). (H) Cellular composition in blood (CD3+, CD19+, Gr-1+), spleen (CD3+, CD19+, Gr-1+), and bone marrow (CD3+, CD19+, Gr-1+, Ter119+, LSK) at week 16 after transplantation.
Analysis of the 4.9-kb deletion containing the HBG2 gene and the HBG2/HBG1 intergenic region
The sgRNA targets identical sequences in the promoters of the HBG1 (Gγ) and HBG2 (Aγ) genes. Therefore, it is expected that a certain percentage of indels will be the monoallelic or biallelic deletion of the 4.9-kb region between the CRISPR cleavage sites, leaving only the HBG1 (Aγ-globin) gene for γ-globin expression (Figure 7A). To analyze this event, we used 2 PCR methods. Quantitative PCR measured the decrease in a signal specific for the intergenic deletion (Figure 7B). A similar method was used by Traxler et al for cells nucleofected with HBG-CRISPR–expressing plasmid.9 In agreement with that study, we detected ∼20% deletions in HUDEP-2 cells transduced with HDAd-HBG-CRISPR. However, we also found ∼2% deletions in CD34+ cells and Lin− cells from β-YAC/CD46 mice in the ex vivo and in vivo HSPC-transduction settings. A second semiquantitative direct PCR method (Figure 7C-D) demonstrated higher deletion levels: 23.4% in HUDEP-2 cells, 3.0% in CD34+ cells, and 5.5%/5.7% in β-YAC/CD46 mice in the ex vivo transduction setting and 1.5%/6.4% in the in vivo transduction setting. The deletion frequency correlated with the CRISPR/Cas9 cleavage efficacy measured by T7E1 assay (Figure 7D). No inversion of the 4.9-kb region was detected by PCR.
Analysis of a 4.9-kb deletion containing the HBG2 gene and the HBG2/HBG1 intergenic region. (A) Diagram showing the HBG1/2 region. The HBG CRISPR/Cas9 cleavage sites are indicated by scissors. Quantitative PCR (qPCR) primers targeting the intergenic sequence between the cleavage sites are indicated (blue half arrows). Another pair of primers binding outside of the deletion region (gray half arrows) was used as an internal control to adjust for differences in template DNA quality. Primers for semiquantitative PCR include the red primers (a 9.9-kb product would indicate no 4.9 kb deletion; a 5.0-kb product would indicate a 4.9-kb deletion), and green primers (a 3.9-kb product would indicate an inversion of the 4.9-kb region). (B) Comparative qPCR to detect the 4.9-kb deletion (blue and gray primers) in (A). HUDEP-2, human CD34+ cells, or βYAC/CD46 Lin− cells (β-YAC-ex vivo) were transduced with HDAd-HBG-CRISPR at a multiplicity of infection of 500, 1000, or 500 vp’s per cell, respectively. For CD34+ cells, a second transduction with HDAd–anti-CRISPR (multiplicity of infection = 1000) was conducted 2 days later. Genomic DNA was isolated at day 4 posttransduction. Furthermore, Lin− cells harvested at week 12 from in vivo HSPC-transduced β-YAC/CD46 mice were analyzed (β-YAC-in vivo). Untransduced samples were used for comparison of qPCR signals. Comparative qPCR was performed in sextuplicates. The signals from the intergenic HBG1/2 region (blue primers) were normalized to corresponding signals from the outside control region (gray primers) in (A). Data were calculated by the 2-ΔΔCt approach (a relative quantification strategy for qPCR data) and are shown as fold change compared with corresponding untransduced cells (taken as 1.0). (C-D) Semiquantitative PCR to detect the 4.9-kb deletion (red primers) or inversion (green primers) in (A). (C) Standard curve for detection of the 4.9-kb deletion. Genomic DNA isolated from untreated wild-type HUDEP-2 cells and a HDAd-HBG-CRISPR–transduced HUDEP-2 clone with a biallelic 4.9-kb deletion (supplemental Materials and methods) were mixed at various ratios and used as a template for PCR. The percentage of the PCR signal corresponding to the deletion is shown above the gel (left panel). Signals were quantified by ImageQuant and used to build a standard curve (right panel). (D) Percentage of the PCR signal corresponding to the 4.9-kb deletion in samples in (B). Two mice for the ex vivo and in vivo HDAd-HBG-CRISPR–transduction settings were used. The lower row of numbers shows the cleavage frequency measured by T7E1 assay in the given samples. Another PCR using the green primers did not show signals, indicating the absence of an inversion of the 4.9-kb region. The primers were validated using a synthesized gBlock as a positive control.
Analysis of a 4.9-kb deletion containing the HBG2 gene and the HBG2/HBG1 intergenic region. (A) Diagram showing the HBG1/2 region. The HBG CRISPR/Cas9 cleavage sites are indicated by scissors. Quantitative PCR (qPCR) primers targeting the intergenic sequence between the cleavage sites are indicated (blue half arrows). Another pair of primers binding outside of the deletion region (gray half arrows) was used as an internal control to adjust for differences in template DNA quality. Primers for semiquantitative PCR include the red primers (a 9.9-kb product would indicate no 4.9 kb deletion; a 5.0-kb product would indicate a 4.9-kb deletion), and green primers (a 3.9-kb product would indicate an inversion of the 4.9-kb region). (B) Comparative qPCR to detect the 4.9-kb deletion (blue and gray primers) in (A). HUDEP-2, human CD34+ cells, or βYAC/CD46 Lin− cells (β-YAC-ex vivo) were transduced with HDAd-HBG-CRISPR at a multiplicity of infection of 500, 1000, or 500 vp’s per cell, respectively. For CD34+ cells, a second transduction with HDAd–anti-CRISPR (multiplicity of infection = 1000) was conducted 2 days later. Genomic DNA was isolated at day 4 posttransduction. Furthermore, Lin− cells harvested at week 12 from in vivo HSPC-transduced β-YAC/CD46 mice were analyzed (β-YAC-in vivo). Untransduced samples were used for comparison of qPCR signals. Comparative qPCR was performed in sextuplicates. The signals from the intergenic HBG1/2 region (blue primers) were normalized to corresponding signals from the outside control region (gray primers) in (A). Data were calculated by the 2-ΔΔCt approach (a relative quantification strategy for qPCR data) and are shown as fold change compared with corresponding untransduced cells (taken as 1.0). (C-D) Semiquantitative PCR to detect the 4.9-kb deletion (red primers) or inversion (green primers) in (A). (C) Standard curve for detection of the 4.9-kb deletion. Genomic DNA isolated from untreated wild-type HUDEP-2 cells and a HDAd-HBG-CRISPR–transduced HUDEP-2 clone with a biallelic 4.9-kb deletion (supplemental Materials and methods) were mixed at various ratios and used as a template for PCR. The percentage of the PCR signal corresponding to the deletion is shown above the gel (left panel). Signals were quantified by ImageQuant and used to build a standard curve (right panel). (D) Percentage of the PCR signal corresponding to the 4.9-kb deletion in samples in (B). Two mice for the ex vivo and in vivo HDAd-HBG-CRISPR–transduction settings were used. The lower row of numbers shows the cleavage frequency measured by T7E1 assay in the given samples. Another PCR using the green primers did not show signals, indicating the absence of an inversion of the 4.9-kb region. The primers were validated using a synthesized gBlock as a positive control.
Discussion
In the current study, we used a nonintegrating HDAd5/35++ vector for HSPC genome editing in vitro and in vivo. Because the CD34+/NSG humanized mouse has limitations in assessing γ-globin reactivation, because it does not support human erythropoiesis, we focused on the β-YAC mouse model. Our study demonstrated that in vitro and in vivo HBG-promoter editing in β-YAC/CD46 HSPCs resulted in reactivation of human γ-globin in erythrocytes of adult animals.
Efficacy of γ-globin reactivation achieved with HDAd-HBG-CRISPR genome editing
The percentage of γ-globin–expressing RBCs in treated mice was ∼20% (Figure 2E) and up 13% (Figure 4G) after ex vivo and in vivo HSPC genome editing, respectively. In secondary recipients of in vivo–transduced β-YAC/CD46 HSPCs, the percentage of γ-globin+ RBCs (Figure 6C) and the target site cleavage efficacy (Figure 6D) were significantly higher (P < .05) than in primary transduced mice. Based on our previous finding that integrating HDAd5/35++ vectors preferentially targets primitive HSCs,20 we speculate that, in the in vivo HDAd-HBG-CRISPR approach, genome editing occurs in self-renewing multilineage HSCs, which becomes more apparent in long-term repopulation studies in secondary recipients.
Notably, in our β-YAC/CD46 mouse model, γ-globin–expressing erythroid cells have no survival advantage. Therefore, our in vivo approach should be more efficient in humanized mouse models of β-thalassemia major and sickle cell disease (SCD), because of the great growth advantage of human γ-globin–expressing erythroid cells.40,41 The latter is supported by transplantation studies in humanized β-thalassemia major mice, in which low-level engraftment of wild-type bone marrow resulted in 90% of circulating donor erythrocytes 5 months after transplantation.42 Studies in a humanized thalassemia major/CD46+/+ model would also answer the question of whether our in vivo approach can achieve therapeutic correction of the disease phenotype.
Reversed β- to γ-globin switch by HBG promoter disruption in β-YAC mice
Developmental control of globin expression is regulated by competition between the corresponding genes for the LCR.1-3 After birth, interaction between the LCR and HBG genes is blocked by proteins, including BCL11A and ZBTB7.19,45 Our study shows, in an in vivo system, that by targeting the binding site of 1 these inhibitors (BCL11A) in the HBG promoter, RBC HBB mRNA levels decrease in the in vivo and ex vivo approach (threefold and fivefold, respectively), whereas HBG mRNA levels greatly increase (10- and 30-fold, respectively). Our data support the critical role for the recently discovered BCL11A binding motif31,32 in the regulation of the β-globin locus in an adequate animal model. Specifically, in SCD, the reverse β- to γ-globin switch has the potential to reduce mutant β-globin chains and to provide γ-globin that can pair with α-globin chains. Our studies showed no detectable γ-globin background expression in nonerythroid cells, most likely due to the strict control by the endogenous LCR.
Future studies have to include HDAd-HBG-CRISPR vectors that more efficiently disrupt the TGACCA motif. With our current vector, only 50% of all indels in the β-YAC/CD46 model and ∼25% in the CD34+/NSG model overlapped with the TGACCA motif (Figure 2E; supplemental Figure 6). In the CD34+/NSG model, the indel frequency was lower, and insertions that would not disrupt the TGACCA motif were more dominant, which could have contributed to the lower degree of γ-globin reactivation compared with the β-YAC/CD46 model. Differences in the type of indels could be due to different DNA damage repair mechanisms in mouse and human cells.11 Another potential way to increase the efficacy of the approach is to include multiple guide RNAs to a given target region in the vector to increase the frequency of knockout in all 4 potential HBG promoter target sites. In support of this, we found, in studies with HDAd-HBG-CRISPR–transduced HUDEP-2 clones, a correlation between the number of indels per clone and the mean fluorescence intensity of reactivated γ-globin (supplemental Figure 16). In this context, it is possible that our γ-globin flow cytometry analysis on RBCs does not include HbF+ cells with only low mean fluorescence intensity due to single indels.
Safety of ex vivo and in vivo HSPC genome editing with HDAd-HBG-CRISPR
Ex vivo transduction of β-YAC/CD46 Lin− cells with HDAd-HBG-CRISPR did not negatively affect their engraftment and differentiation potential. However, although mouse HSCs are less sensitive to DNA double-strand breaks (DSBs),11 quiescent human HSCs are sensitized to apoptosis after DSB-inducing irradiation.46 In this context, CRISPR/Cas9-mediated DSBs in human CD34+ cells have been associated with diminished potential to engraft in NSG mice due to prolonged expression of CRISPR/Cas9 after plasmid transfection and HDAd5/35++ transduction.12,47 Several approaches can be used to address this problem, including the timed expression of anti-CRISPR peptides36 or sgRNAs against Cas9 to control the duration of CRISPR/Cas9 activity.
Our PCR studies detected, at a low frequency, a 4.9-kb deletion resulting from the CRISPR/Cas9 cleavage in the HBG1 and HBG2 promoters in ex vivo–transduced CD34+ cells and Lin− cells isolated from β-YAC/CD46 mice (that underwent the ex vivo and in vivo HSPC-transduction approach). The 4.9-kb deletion containing the HBG2 (Gγ) gene could also have contributed to the pronounced increase in Aγ chains in HDAd-HBG-CRISPR–edited mice (Figures 5C, 6G). Considering the study by Traxler et al9 and our data in Figure 7D, the frequency of the 4.9-kb deletion appears to depend on the overall cleavage frequency, in a CRISPR/Cas9 dose-dependent manner, rather than on the CRISPR/Cas9 delivery method (electroporation of CRISPR plasmids vs helper-dependent adenoviral vectors). Although it does not challenge the principle that HBG-CRISPR results in the reactivation of γ-globin, the fact that this large deletion occurs should be considered in the potential clinical translation of the approach, because it is not known whether the intergenic HBG2-HBG1 region bears critical functional elements.
To our knowledge, this is the first study demonstrating in vivo HSPC editing after intravenous injection of a CRISPR/Cas9 vector. We recently showed that intravenous injection of HDAd5/35++ vectors does not result in transgene expression in tissues other than mobilized HSPCs, PBMCs, and a specific set of splenic macrophages in CD46-transgenic mice.20 This is in agreement with our early studies in baboons using intravenously injected first-generation CD46-targeting Ad5/35 and Ad5/11 vectors.48 A potential explanation is that CD46 receptor density and accessibility are not sufficiently high on nonhematopoietic tissues to allow for efficient viral transduction.20,49 Intravenous injection of HDAd vectors (as well as other viral vectors) is associated with the release of proinflammatory cytokines,50,51 which can be blocked efficiently by pretreatment with glucocorticoids52 or vector dose fractionation.53 Good safety profiles of intravenously injected oncolytic adenoviruses have been documented in dozens of clinical trials, including a trial with a CD46-targeting oncolytic adenovirus.54
Although our in vivo HSPC-transduction approach would simplify the thalassemia gene, the safety of in vivo CRISPR/Cas9 HSPC genome editing first has to be clearly documented in stringent long-term studies in nonhuman primates. Furthermore, considering the recent development of xenograft models that have greatly improved human erythropoiesis with faithful hemoglobin expression,55,56 more safety and efficacy studies in humanized mice have to be conducted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Zhinan Liu for help with the indel sequence analysis.
This work was supported by National Institutes of Health, National Cancer Institute grant R21CA193077 and National Institutes of Health, National Heart, Lung, and Blood Institute grant R01HL128288, the Wings of Karen Foundation, the Marsha Rivkin Foundation, and Grants4Targets from Bayer US LLC (A.L.). N.P. was supported by Cooley’s Anemia Foundation.
Authorship
Contribution: A.L. provided the conceptual framework for the study and wrote the manuscript; C.L., N.P., and A.L. designed the experiments; C.L., N.P., P.S., S.G., H.W., C.V., J.K., and C.K. performed the experiments; R.D.H. provided critical support for sequencing; and G.S. provided β-YAC mice and critical comments on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: André Lieber, University of Washington, Box 357720, Seattle, WA 98195; e-mail: lieber00@uw.edu.
REFERENCES
Author notes
C.L. and N.P. contributed equally to this work.


![Figure 1. Studies with human CD34+ cells. (A) Schematic structure of globin locus with localization of the sgRNA in the promoters of the γ-globin genes, the CRISPR/Cas9 cleavage site (arrowhead), the 13-bp (−114 to −102) HPFH deletion (box), and the BCL11A-binding motif (“TGACCA” bold sequence). Note that only the distal TGACCA motif (red) within the indicated sequence is critical for γ-globin silencing.32 The CRISPR/Cas9 cleavage site is located 2 bp upstream of the BCL11A binding motif. ε, Gγ, Aγ, δ, and β are globin genes. (B) HDAd-HBG-CRISPR vector structure. The sgRNA gene is transcribed by PolIII from the U6 promoter, and the spCas9 gene is under the control of the EF1α promoter. Cas9 expression is controlled by miR-183-5p and miR-218-5p, which suppress Cas9 expression in HDAd producer cells but do not negatively affect Cas9 expression in CD34+ cells.35 The corresponding microRNA target sites (miR-T) were embedded into a 3′untranslated region (3′UTR). (C) Experimental design. CD34+ cells were transduced with HDAd-HBG-CRISPR at a multiplicity of infection of 2000 vp’s per cell. Two days later, cells were infected with HDAd–anti-CRISPR36 at the same multiplicity of infection to terminate CRISPR/Cas9 activity and, thus, minimize cytotoxicity to HSPCs. (The 2 anti-CRISPR peptides [AcrII4 and AcrII2] expressed from this vector are capable of binding to the CRISPR/Cas9 complex, thus blocking its activity.57) Sixteen hours after the last infection, CD34+ cells were transplanted into irradiated NSG mice. Untransduced cells were incubated for 2 days + 16 hours in the same medium as transduced cells. At week 10 after transplantation, human CD45+ cells were isolated from the bone marrow by magnetic-activated cell sorting and subjected to T7E1 assay and erythroid in vitro differentiation for globin analysis by flow cytometry and HPLC. (D) Target site cleavage frequency (measured by T7E1 assay) in human CD45+ cells isolated from bone marrow at week 10 after transplantation. Each lane is an individual mouse. (E) γ-Globin flow cytometry of cells after erythroid in vitro differentiation. (F) Ratio of γ-globin to adult α- or β-globin measured by HPLC after erythroid differentiation. n = 3. HS, DNase I hypersensitivity sites; untr, mice transplanted with untransduced CD34+ cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/26/10.1182_blood-2018-03-838540/4/m_blood838540f1.jpeg?Expires=1765903390&Signature=SlF2lVW7BhiOfmXj~NvfCZF2vLseF1Z1q4K4Wdyj2JjyOZZtLSSUfO2~FogtfbGMUw1HcPERXFEoxJapqdEnH05IO2nMUtcr16o5Zmv8ueMz3DzDJQuVLzKhaWcOjazfSj-2dqfqqhVhT4dispcGb-bKV2mAJYXwwf8iGc0xLCSJ6wX7~uZJtF47M7O4K4Jc6A~ScDoZnQ3RAG80bUK9VL1PJDm4VUtxRSPN3ipYzrbXhlMt58IkqR2a6p3k~LokD~75GP7eeUiqr0Fy4Oc7gDCe~Tg55LkfJbbZRUIe2kY1jnzVflIFyEhMp1eJc48A6au-mvRxyQEnJT2umVjjWg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal