Key Points
We conducted the first exome-wide association study between germ line variant genotype and survival outcomes after unrelated-donor BMT.
A number of novel genes were found to significantly affect survival outcomes.
Abstract
Although survival outcomes have significantly improved, up to 40% of patients die within 1 year of HLA-matched unrelated-donor blood and marrow transplantation (BMT). To identify non-HLA genetic contributors to mortality after BMT, we performed the first exome-wide association study in the DISCOVeRY-BMT cohorts using the Illumina HumanExome BeadChip. This study includes 2473 patients with acute myeloid leukemia, acute lymphoblastic leukemia, or myelodysplastic syndrome and 2221 10/10 HLA-matched donors treated from 2000 to 2011. Single-variant and gene-level analyses were performed on overall survival (OS), transplantation-related mortality (TRM), and disease-related mortality (DRM). Genotype mismatches between recipients and donors in a rare nonsynonymous variant of testis-expressed gene TEX38 significantly increased risk of TRM, which was more dramatic when either the recipient or donor was female. Using the SKAT-O test to evaluate gene-level effects, variant genotypes of OR51D1 in recipients were significantly associated with OS and TRM. In donors, 4 (ALPP, EMID1, SLC44A5, LRP1), 1 (HHAT), and 2 genes (LYZL4, NT5E) were significantly associated with OS, TRM, and DRM, respectively. Inspection of NT5E crystal structures showed 4 of the associated variants affected the enzyme structure and likely decreased the catalytic efficiency of the enzyme. Further confirmation of these findings and additional functional studies may provide individualized risk prediction and prognosis, as well as alternative donor selection strategies.
Introduction
Blood and marrow transplantation (BMT) is an effective treatment for many life-threatening malignant hematologic diseases. Survival after BMT has increased dramatically over the past 2 decades, as a result of improvements in patient selection, high-resolution HLA typing, better supportive care, and infection prophylaxis.1-3 The most established genetic predictors of BMT survival reside in the major histocompatibility complex (MHC) region, which encodes the HLAs. To improve survival, the current recommendation is to select a donor who matches the recipient in 4 HLA genomic loci (HLA-A, -B, -C, and -DRB1) as well as other characteristics, including HLA-DP typing, sex, blood type, and cytomegalovirus status.4-7 Numerous candidate gene studies have investigated non-HLA genomic loci that are associated with survival after allogeneic BMT.8 However, these studies have generated inconsistent results, likely because of sample size limitations and heterogeneity of disease and disease status, recipient-donor relationship, degree of HLA match, and other factors.8 As part of the DISCOVeRY-BMT study (Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT),9,10 we conducted the first exome-wide association study of mortality after BMT. Exome-wide association studies analyze functional coding variants, including those that are rare in the population, under the hypothesis that rare genetic variants may convey larger effects than common variants.11,12 Our study is the first analysis of the rare variant contribution to BMT survival outcomes, from which we identified both missense variants and novel genes associated with patient survival after HLA-matched unrelated-donor (URD) allogeneic BMT.
Methods
Study population
All participants were from the DISCOVeRY-BMT study.9,10 DISCOVeRY-BMT consists of 2 well-characterized cohorts (cohort 1 and cohort 2; Table 1). Both cohorts include patients who were treated with URD BMT for ALL, AML, and MDS and their 8/8 HLA-matched (high-resolution HLA-A, -B, -C, and -DRB1 loci) unrelated donors. Cohort 1 includes patients who underwent 10/10 HLA-matched (high-resolution HLA-A, -B, -C, -DRB1, and -DQB1 loci) URD BMT from 2000 to 2008 and reported to the Center for International Blood and Marrow Transplant Research, whereas cohort 2 includes patients who underwent 10/10 HLA-matched URD BMT from 2009 to 2011 or 8/8 (but <10/10) HLA-matched URD BMT from 2000 to 2011 and reported to Center for International Blood and Marrow Transplant Research. The exome-wide association study was restricted to patients who received a 10/10 HLA match in cohort 1 or 2. Causes of death were adjudicated by an expert panel as previously described.9 All patients and donors provided written informed consent for their clinical data to be used for research purposes and were not compensated for their participation. This study was reviewed and approved by the Roswell Park Cancer Institute Institutional Review Board. All patient data were deidentified.
Characteristics of the DISCOVeRY-BMT cohort
| Patient characteristics . | N (%) . | P* . | |
|---|---|---|---|
| Cohort 1 (n = 1970) . | Cohort 2 (n = 503) . | ||
| Age, y | <.01 | ||
| Mean | 43.0 | 46.7 | |
| Range | 0.45-74.5 | 0-78 | |
| Donor age, y | <.01 | ||
| Mean | 34.2 | 31.6 | |
| Range | 18.4-61.2 | 18.2-60 | |
| Sex | NS | ||
| Female | 858 (44) | 218 (43) | |
| Male | 1112 (56) | 285 (57) | |
| Donor sex | <.01 | ||
| Female | 564 (32) | 118 (25) | |
| Male | 1177 (68) | 362 (75) | |
| Disease | <.01 | ||
| ALL | 442 (22) | 41 (8) | |
| AML | 1196 (61) | 327 (65) | |
| MDS | 332 (17) | 135 (27) | |
| ALL disease status | NS | ||
| Early | 362 (82) | 33 (80) | |
| Advanced | 80 (18) | 8 (20) | |
| AML disease status | .01 | ||
| Early | 843 (70) | 254 (78) | |
| Advanced | 353 (30) | 73 (22) | |
| MDS disease status | <.01 | ||
| Early | 221 (67) | 46 (34) | |
| Advanced | 111 (33) | 89 (66) | |
| Graft source | <.01 | ||
| Bone marrow | 694 (35) | 114 (23) | |
| Peripheral blood | 1276 (65) | 389 (77) | |
| Patient characteristics . | N (%) . | P* . | |
|---|---|---|---|
| Cohort 1 (n = 1970) . | Cohort 2 (n = 503) . | ||
| Age, y | <.01 | ||
| Mean | 43.0 | 46.7 | |
| Range | 0.45-74.5 | 0-78 | |
| Donor age, y | <.01 | ||
| Mean | 34.2 | 31.6 | |
| Range | 18.4-61.2 | 18.2-60 | |
| Sex | NS | ||
| Female | 858 (44) | 218 (43) | |
| Male | 1112 (56) | 285 (57) | |
| Donor sex | <.01 | ||
| Female | 564 (32) | 118 (25) | |
| Male | 1177 (68) | 362 (75) | |
| Disease | <.01 | ||
| ALL | 442 (22) | 41 (8) | |
| AML | 1196 (61) | 327 (65) | |
| MDS | 332 (17) | 135 (27) | |
| ALL disease status | NS | ||
| Early | 362 (82) | 33 (80) | |
| Advanced | 80 (18) | 8 (20) | |
| AML disease status | .01 | ||
| Early | 843 (70) | 254 (78) | |
| Advanced | 353 (30) | 73 (22) | |
| MDS disease status | <.01 | ||
| Early | 221 (67) | 46 (34) | |
| Advanced | 111 (33) | 89 (66) | |
| Graft source | <.01 | ||
| Bone marrow | 694 (35) | 114 (23) | |
| Peripheral blood | 1276 (65) | 389 (77) | |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; NS, not significant.
Kolmogorov-Smirnov test was used to compare the age distribution between cohorts. Fisher’s exact test and χ2 test weres used to compare binary and multicategorical data between the 2 cohorts.
Genotyping and quality control
All samples were genotyped using Illumina HumanExome-12 v1.1 BeadChip. We used the Optimal Sample Assignment Tool to randomly assign samples to plates for genotyping to eliminate batch effects and any potential confounding factors.13 Samples and variants that were successfully genotyped underwent stringent sample-level and variant-level quality control (QC). In sample-level QC, samples were removed if the missing rate was ≥2%, if the typed and reported sex did not match, if there were abnormal inbreeding coefficients, or if cryptic relatedness was present. Population outliers were removed using EIGENSTRAT.14 Because of the low number of non-European populations in our study cohort, our analyses were carried out only in recipients of European descent and their donors. After sample-level QC, 1970 recipients and 1741 donors were retained in cohort 1 and 503 recipients and 480 donors were retained in cohort 2. In variant-level QC, variants were removed if the missing rate was ≥2%, if there was ≥1 Mendelian error in HapMap trios included in our genotyping, or if variants violated Hardy-Weinberg equilibrium (P < 1 × 10−6) or were discordant between duplicate samples. In total, 240 653 variants in recipients and 240 640 variants in donors of cohort 1 successfully passed QC, and 240 603 variants in recipients and 240 573 variants in donors of cohort 2 successfully passed QC. Cluster plots for all variants reported herein were manually inspected for quality.
Statistical analysis
Because the 2 cohorts differed on several patient, disease, and treatment characteristics, we used a 2-stage study design (supplemental Figure 1, available on the Blood Web site).
Single-variant analysis
The likelihood ratio test (LRT) was used to analyze single-variant association between survival outcomes of recipients (overall survival [OS]/treatment-related mortality [TRM]/disease-related mortality [DRM]) and the variant genotypes of either recipients or corresponding donors while controlling for additional covariates of recipients: age at BMT, disease diagnosis (AML/ALL/MDS), disease status at BMT, cell source (peripheral blood/marrow), and all principal components deemed significant by the Tracy-Widom statistic derived from EIGENSTRAT. Singleton variants and nonautosome variants were eliminated from the single-variant association analysis. We used cumulative incidence functions to handle the competing risks of TRM and DRM in association analysis. We also used LRT to evaluate the association between recipients’ survival outcomes and the recipient-donor allele mismatches for each variant, defined as the absolute difference in allele dosage between each recipient-donor pair. The survival and cmprsk packages in R software were used to perform single-variant analysis within each cohort, and the meta-analysis of both cohort 1 and cohort 2 was performed using a fixed-effects model in METAL.15 For each outcome, variants with a suggestive association (P < 1 × 10−3) in cohort 1 were entered into the fixed-effects model meta-analysis performed using METAL.15 Significance level was determined using a Bonferroni correction for the effective number of independent tests after adjusting for the correlation between variants,16,17 which yielded Pmeta ≤ 1.32 × 10−6 to reach exome-wide significance. To evaluate whether sex modified the association between TRM and recipient-donor allele mismatches in rs200092801, we added an interaction term between the sex indicator variable (for male-male recipient donor pairs) and the allele mismatches in rs200092801 to the competing risk model and used an LRT to assess the significance of the interaction term.
Gene-based analysis
The optimal sequence kernel association (SKAT-O) test18 was used to analyze the gene-level association between recipients’ survival outcomes (OS/TRM/DRM) and the genotypes of either recipients or the corresponding donors using the same covariates as the single-variant analysis. In gene-level analysis, we adopted a cause-specific hazard model to handle the competing risk situation in association analysis of TRM and DRM. The skatMeta and survival packages in R software were used for running the SKAT-O tests. We removed variants with minor allele frequency >1% from the corresponding cohorts and only kept missense and nonsense variants for the analysis, which resulted in >12 000 genes tested in recipients and donors. In the discovery stage, we performed SKAT-O tests on genes with ≥2 variants in cohort 1. The genes with P < 1 × 10−3 were further evaluated in a meta-analysis that integrated genotypes from both cohort 1 and cohort 2. We set overall exome-wide significance at Pmeta < 4.10 × 10−6 after Bonferroni correction for the total number of genes tested.
MHC binding prediction
MHC binding affinity was predicted across all possible 8- to 15-mer peptides surrounding the rs200092801 variant in TEX38 and the corresponding wild-type peptides using NetMHCpan 4.0.19 These tiled peptides were analyzed for their binding affinities (IC50 in nM units) to each class I HLA allele. An IC50 value of <150 nM was considered a predicted strong binder, and IC50 >500 nM was considered a nonbinder.
Crystal structure analysis
The crystal structures of NT5E were obtained from the Protein Data Bank (Protein Data Bank codes: 4H2G [open state], 4H2I [closed state]). Visualization was performed using PyMOL Molecular Graphics System, version 1.8 (Schrödinger, https://www.schrodinger.com/suites/pymol).
Kaplan-Meier plots
Kaplan-Meier (KM) plots were used to show the impact of the gene on survival outcomes. For the KM plots, we assumed variant impact on survival was exchangeable, with each variant contributing equally in magnitude and direction. Therefore, we only generated the KM plots when the assumption of directionality was not violated based on the variant weights from SKAT-O.
Results
Single-variant association analyses
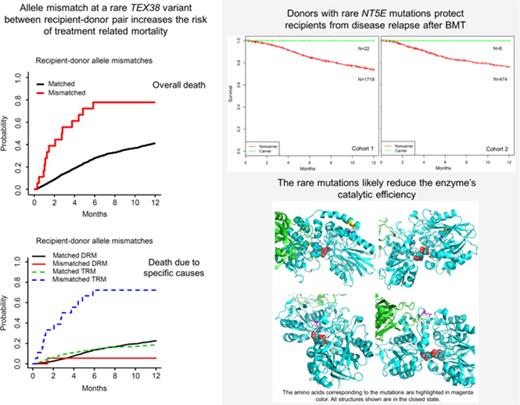
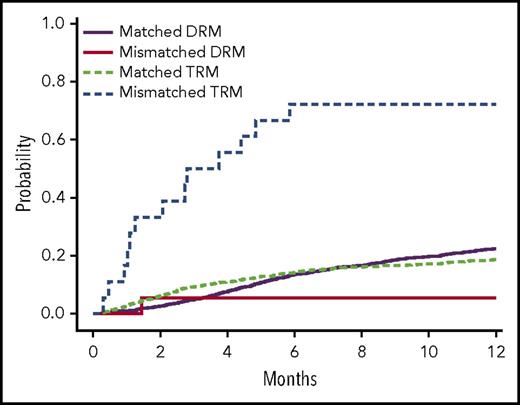
The single-variant association analyses of recipient (only) genotypes and donor (only) genotypes did not yield any statistically significant variants. In the analysis of recipient-donor variant allele mismatching, only 1 variant in TEX38 (exm56988, rs200092801) was statistically significant in the single-variant analyses (Table 2; Figure 1; supplemental Figure 2). TRM in recipients was found to be significantly associated with the allele mismatches between recipients and donors of rs200092801 (Pmeta = 3.51 × 10−7). Recipients whose genotypes differed from their donors’ were much more likely to die as a result of TRM than those who had genotypes that were identical to their donors’, but there was no impact of this variant on DRM (supplemental Table 1; supplemental Figure 3). This association between TRM and allele mismatches at rs200092801 likely resulted from the consistent trend of increasing TRM risk by the minor allele of rs200092801 in both recipient and donor genotypes (supplemental Table 1; supplemental Figure 3). rs200092801 is a rare nonsynonymous variant and was observed only in recipients of cohort 1 with minor allele frequency of 0.3%. TEX38 is highly and preferentially expressed in the testes, but its function is not yet understood. Given the strong expression of TEX38 in the testes, the 4 combinations of recipient-donor sex (male donor to female recipient [M→F], male donor to male recipient [M→M], female donor to male recipient [F→M], and female donor to female recipient [F→F]) were assessed for an interaction with rs200092801 for TRM. Recipient-donor pairs were stratified based on these 4 combinations, and variant association was analyzed with a competing risk model. Recipient hazard of death resulting from TRM was considerably higher when either the recipient or donor was female and recipient and donor were mismatched at rs200092801, compared with male recipients with male donors who were mismatched at this variant (Figure 2). Specifically, among recipients who were mismatched with their donors at rs200092801, every female recipient and every recipient with a female donor died as a result of TRM, compared with 44% patients in the M→M group who died as a result of TRM. The median survival times for recipient-donor pairs mismatched at rs200092801 for F→F, F→M, and M→F were 1.1, 0.9, and 4.4 months, respectively; the median survival time for M→M was 5.9 months. The interaction between recipient-donor sex combination and recipient-donor allele mismatch at rs200092801 were tested in the competing risk model, and recipient-donor sex was found to significantly modify the association between TRM and recipient-donor allele mismatches at rs200092801 (Pinteraction = 4.6 × 10−3).
The variant in TEX38 whose allele mismatches contribute to TRM in recipients
| Variant . | Chromosome . | Position . | Alleles (ref/alt) . | rsID . | AA change* . | Impact† . | Minor allele . | MAF‡ . | Allele mismatch frequency§ . | Population MAF‖ . |
|---|---|---|---|---|---|---|---|---|---|---|
| exm56988 | 1 | 47139039 | C/T | rs200092801 | P178S | Tolerated | T | 3.05E−03 | 5.17E−03 | 1.60E−03 |
| Variant . | Chromosome . | Position . | Alleles (ref/alt) . | rsID . | AA change* . | Impact† . | Minor allele . | MAF‡ . | Allele mismatch frequency§ . | Population MAF‖ . |
|---|---|---|---|---|---|---|---|---|---|---|
| exm56988 | 1 | 47139039 | C/T | rs200092801 | P178S | Tolerated | T | 3.05E−03 | 5.17E−03 | 1.60E−03 |
MAF, minor allele frequency.
The amino acid changes are based on transcript NM_001145474.
The MAF in recipients of cohort 1. The minor allele was not observed in recipients or donors of cohort 2.
The frequency of allele mismatch in recipient-donor pairs in cohort 1. This was calculated as the sum of absolute difference in allele dosage between all recipient-donor pairs divided by 2 times the number of recipient-donor pairs.
The MAF in the Exome Aggregation Consortium58 non-Finnish European population.
Estimated cumulative incidence curves with TRM and DRM as competing events for recipients with donor genotypes with 1 allele mismatch and no mismatch of rs200092801 in cohort 1. Two allele mismatches of this variant between recipients and donors were not observed.
Estimated cumulative incidence curves with TRM and DRM as competing events for recipients with donor genotypes with 1 allele mismatch and no mismatch of rs200092801 in cohort 1. Two allele mismatches of this variant between recipients and donors were not observed.
Estimated cumulative incidence curves for different combinations of recipient and donor sex with TRM and DRM as competing events for recipients with donor genotypes with 1 allele mismatch and no mismatch of rs200092801 in cohort 1. Two allele mismatches of this variant between recipients and donors were not observed.
Estimated cumulative incidence curves for different combinations of recipient and donor sex with TRM and DRM as competing events for recipients with donor genotypes with 1 allele mismatch and no mismatch of rs200092801 in cohort 1. Two allele mismatches of this variant between recipients and donors were not observed.
Suspecting TEX38 as a male testis antigen, we further explored whether rs200092801 could affect the recognition of the antigen by HLA class I molecules. rs200092801 causes an amino acid change (P178S) in TEX38 protein. Interestingly, we identified 3 peptides containing the mutant amino acid S (mutant peptides) that were predicted to strongly bind with various HLA class I alleles, whereas the corresponding peptides containing the wild-type amino acid P (wild-type peptides) had no binding (supplemental Table 2). Therefore, it is possible that the C to T nucleotide change of rs200092801 in TEX38 results in a mutant peptide that can be presented by MHC class I molecules to immune cells to trigger downstream immune response and TRM. Among the 18 recipient-donor pairs with allele mismatches at rs200092801, there were 11 pairs carrying the HLA class I alleles that were predicted to strongly bind with the mutant peptides but not the wild-type peptides. Notably, recipients in 9 (82%) of these 11 pairs died as a result of TRM, compared with 4 TRM events (57%) in the 7 remaining pairs, which suggests a positive link between TRM risk and rs200092801-specific strong binding of TEX38 peptide with MHC class I molecules.
Gene-level association analyses
Recipient gene-level associations with OS, TRM, and DRM after BMT.
One gene, OR51D1, which encodes an olfactory receptor (OR), was significantly associated with OS (Pmeta = 9.48 × 10−7). Six nonsynonymous variants in OR51D1 contributed to the association signal (Table 3); all are rare in the DISCOVeRY-BMT cohorts and in the general European population. Importantly, OR51D1 was also the only gene significantly associated with TRM (Pmeta = 1.05 × 10−6), with the same 6 variants contributing to the associations signal. This gene was not associated with DRM (Pmeta = .07), which means the genetic contribution of OR51D1 to TRM translates to a significant effect on OS. An advantage of SKAT-O is that it allows individual variants to have different directions and magnitudes of effect. In association analysis of OS, the minor alleles of 5 of the 6 variants contributed to better survival, whereas in association analysis of TRM, the minor alleles of all 6 variants contributed to better survival (supplemental Table 3). In an intuitive visualization of the impact of the gene on survival outcomes, supplemental Figure 4 compares recipients who carried a minor allele at any of the 6 variants and those who did not. There was a protective effect of OR51D1 variants in recipients that reduced the risk of TRM; none of the recipients carrying these 6 minor alleles died as a result of TRM, although there were some deaths attributable to DRM. We did not find any recipient genes to be significantly associated with DRM.
The 6 variants in OR51D1 that contribute to the gene-level association with OS and TRM in recipients
| Variant . | Chromosome . | Position . | Alleles (ref/alt) . | rsID . | AA change* . | Impact† . | Minor allele . | MAF . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Cohort 1 . | Cohort 2 . | Population‡ . | ||||||||
| exm881159 | 11 | 4661190 | C/G | rs138224979 | T57S | Tolerated | G | 2.54E−04 | NA | 4.50E−05 |
| exm881165 | 11 | 4661280 | C/T | rs148606808 | S87F | Deleterious | T | 3.81E−03 | 4.97E−03 | 2.40E−03 |
| exm881174 | 11 | 4661373 | A/C | rs141786655 | H118P | Deleterious | C | 5.08E−04 | 9.94E−04 | 3.00E−04 |
| exm881176 | 11 | 4661424 | G/A | rs61745314 | R135H | Deleterious | A | 2.54E−04 | 9.94E−04 | 1.50E−05 |
| exm881178 | 11 | 4661451 | G/A | rs200394876 | R144H | Tolerated | A | 1.02E−03 | NA | 2.00E−04 |
| exm881179 | 11 | 4661457 | C/T | rs149135276 | A146V | Deleterious | T | 7.61E−04 | 9.94E−04 | 1.00E−03 |
| Variant . | Chromosome . | Position . | Alleles (ref/alt) . | rsID . | AA change* . | Impact† . | Minor allele . | MAF . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Cohort 1 . | Cohort 2 . | Population‡ . | ||||||||
| exm881159 | 11 | 4661190 | C/G | rs138224979 | T57S | Tolerated | G | 2.54E−04 | NA | 4.50E−05 |
| exm881165 | 11 | 4661280 | C/T | rs148606808 | S87F | Deleterious | T | 3.81E−03 | 4.97E−03 | 2.40E−03 |
| exm881174 | 11 | 4661373 | A/C | rs141786655 | H118P | Deleterious | C | 5.08E−04 | 9.94E−04 | 3.00E−04 |
| exm881176 | 11 | 4661424 | G/A | rs61745314 | R135H | Deleterious | A | 2.54E−04 | 9.94E−04 | 1.50E−05 |
| exm881178 | 11 | 4661451 | G/A | rs200394876 | R144H | Tolerated | A | 1.02E−03 | NA | 2.00E−04 |
| exm881179 | 11 | 4661457 | C/T | rs149135276 | A146V | Deleterious | T | 7.61E−04 | 9.94E−04 | 1.00E−03 |
NA, not applicable.
The amino acid changes are based on transcript NM_001004751.
The MAF in the Exome Aggregation Consortium58 non-Finnish European population.
Donor gene-level associations with OS after BMT.
Four genes in donors were significantly associated with OS: ALPP, EMID1, SLC44A5, and LRP1 (Table 4; supplemental Tables 4 and 5). ALPP encodes an alkaline phosphatase, and 3 ALPP variants contributed to the gene-level association. All 14 recipients whose donors carried the minor alleles of these 3 variants survived the first year after BMT (supplemental Figure 5). Two donor variants in EMID1 affected recipient OS; 1 was protective, and the other was detrimental (supplemental Table 5). SLC44A5 is a member of the solute carrier gene family, and 2 donor variants in this gene contributed to its association with OS. Recipients whose donors carried the minor alleles of the 2 variants were found to have worse survival than recipients whose donors were noncarriers (supplemental Figure 6). LRP1 (low-density lipoprotein receptor-related protein 1) encodes an endocytic receptor involved in several cellular processes, including intracellular signaling, lipid homeostasis, and clearance of apoptotic cells. Within LRP1, 27 variants contributed to the association of the gene with OS; however, the effects of the individual variants included some that were protective and some that were risk conveying (supplemental Table 5). None of these 4 genes were significantly associated with TRM or DRM.
The genes significantly associated with survival outcomes in donors
| Outcome . | Gene . | Chromosome . | Pmeta . | No. of variants* . |
|---|---|---|---|---|
| OS | ALPP | 2 | 1.05E−06 | 3 |
| EMID1 | 22 | 1.05E−06 | 2 | |
| SLC44A5 | 1 | 1.05E−06 | 2 | |
| LRP1 | 12 | 2.86E−06 | 27 | |
| TRM | HHAT | 1 | 9.34E−07 | 6 |
| DRM | LYZL4 | 3 | 1.05E−06 | 3 |
| NT5E | 6 | 1.05E−06 | 6 |
| Outcome . | Gene . | Chromosome . | Pmeta . | No. of variants* . |
|---|---|---|---|---|
| OS | ALPP | 2 | 1.05E−06 | 3 |
| EMID1 | 22 | 1.05E−06 | 2 | |
| SLC44A5 | 1 | 1.05E−06 | 2 | |
| LRP1 | 12 | 2.86E−06 | 27 | |
| TRM | HHAT | 1 | 9.34E−07 | 6 |
| DRM | LYZL4 | 3 | 1.05E−06 | 3 |
| NT5E | 6 | 1.05E−06 | 6 |
The number of single variants included in the meta SKAT-O test.
Donor gene-level associations with TRM and DRM after BMT.
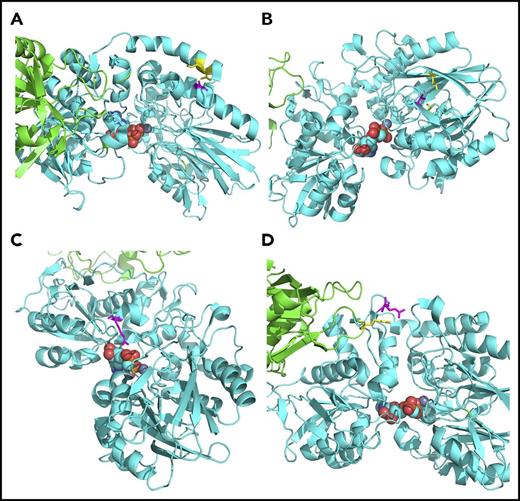
The gene HHAT was significantly associated with TRM, whereas 2 other genes, LYZL4 and NT5E, were significantly associated with DRM (Table 4; supplemental Tables 6-9). HHAT (hedgehog acyltransferase) encodes an enzyme that catalyzes N-terminal palmitoylation of sonic hedgehog (SHH) and is required for SHH signaling. SHH is known to affect T-cell proliferation and maturation and maintain survival and growth of CML progenitor cells.20,21 Six HHAT variants contributed to its association with TRM; 5 of the variants had a protective effect in both cohorts, with 1 variant showing an inconsistent direction of effect between cohorts (supplemental Tables 6 and 7). LYZL4 (lysozyme-like 4) belongs to a family of lysozyme-like genes, and lysozymes play a mainly protective role in host defense. Three LYZL4 variants contributed to the association of the gene with DRM; 2 variants increased risk of DRM, and 1 variant was protective (supplemental Tables 8 and 9). The encoded protein of NT5E, ecto-5′-nucleotidase or CD73, catalyzes the conversion of extracellular nucleotides to membrane-permeable nucleosides. A total of 6 variants contributed to the association of NT5E with DRM, and all exerted a protective effect (supplemental Table 9). None of the recipients whose donors carried the minor alleles of these 6 variants died as a result of DRM, although some of them died as a result of TRM (supplemental Figure 7). We observed the same trend in both patients with ALL and those with AML/MDS (supplemental Figures 8 and 9). NT5E is involved in metabolism of purine analogs, such as fludarabine (Flu), mycophenolate mofetil (MMF), and azathioprine. Flu can be used as part of the conditioning regimen, and MMF and azathioprine are drugs for posttransplantation immunosuppression. We first speculated that NT5E might influence DRM through its interaction with Flu or MMF. However, because there were only 28 recipients with corresponding donors carrying the NT5E variants, and none of them died as a result of disease in the first year after transplantation, we were unable to test for differences in DRM between those receiving the drug or not (data not shown). In contrast, NT5E is the enzyme for converting extracellular adenosine monophosphate to adenosine, which suppresses antitumor T-cell responses and therefore promotes tumor initiation and progression.22-26 Crystal structures of NT5E revealed that the enzyme forms a homodimer and switches between the open and closed conformations in the catalysis process27 (supplemental Figure 10). Four of the 6 variants in donors that protected patients from death resulting from DRM after BMT were predicted to be deleterious (supplemental Table 8). By inspecting the NT5E crystal structures, we found 4 of these mutations to have an impact on the enzyme structure (Figure 3), which likely decreased the catalytic efficiency of the enzyme and led to enhanced antitumor T-cell responses and lower risk of DRM.
The NT5E variants in the crystal structure of the enzyme. (A) NT5E A62 (magenta) directly interacts with I320 and K321 (yellow) in the α-helix that connects the 2 protein domains and works as a hinge for the conformational change between the open and closed states. The A62S variant is predicted to impede the conformational change of the enzyme from the open state to the closed state. (B) V278 (magenta), interacting with L290 and V240 (yellow), is buried within the core of the N-terminal domain. The V278I variant is predicted to affect the folding of this domain and the conformation of the catalytic pocket. (C) The side chain of R354 (magenta) forms a hydrogen bond with adenosine. The R354C variant is predicted to disrupt the binding of adenosine.27 (D) R401 (magenta) interacts with R480 (yellow) from the other chain in the closed state. The R401C variant is predicted to impede the NT5E dimerization and the conformational change of the enzyme from the open state to the closed state. All structures shown are in the closed state. The substrate adenosine is displayed in the sphere model, and the highlighted amino acids are displayed in the stick model.
The NT5E variants in the crystal structure of the enzyme. (A) NT5E A62 (magenta) directly interacts with I320 and K321 (yellow) in the α-helix that connects the 2 protein domains and works as a hinge for the conformational change between the open and closed states. The A62S variant is predicted to impede the conformational change of the enzyme from the open state to the closed state. (B) V278 (magenta), interacting with L290 and V240 (yellow), is buried within the core of the N-terminal domain. The V278I variant is predicted to affect the folding of this domain and the conformation of the catalytic pocket. (C) The side chain of R354 (magenta) forms a hydrogen bond with adenosine. The R354C variant is predicted to disrupt the binding of adenosine.27 (D) R401 (magenta) interacts with R480 (yellow) from the other chain in the closed state. The R401C variant is predicted to impede the NT5E dimerization and the conformational change of the enzyme from the open state to the closed state. All structures shown are in the closed state. The substrate adenosine is displayed in the sphere model, and the highlighted amino acids are displayed in the stick model.
Discussion
This study on the contribution of rare variants to BMT survival outcomes has revealed an interesting sex-specific single-variant association and a number of novel genes. Genotype mismatches between recipients and donors in a rare variant of TEX38 (rs200092801) significantly increased the risk of TRM. The increase of TRM risk was less dramatic when the recipients and the corresponding donors were both male. TEX38 is highly expressed in the testes with unknown function. The testes, which were previously thought to be insulated from the immune system by the Sertoli cell barrier, were recently found to be connected with immune system.28 In this groundbreaking study, the authors demonstrated in mice that there are nonsequestered meiotic germ cell antigens (NS-MGCAs) that can pass through the Sertoli cell barrier and then be recognized by antigen-specific regulatory T cells to maintain systematic tolerance to NS-MGCAs.28 The better tolerance of genotype mismatches at rs200092801 when both recipients and donors were male may therefore be related to the systematic tolerance for NS-MGCAs in males. We further investigated why genotype mismatches at rs200092801 could affect TRM, which is a composite of mainly immune-related causes. We speculated this could happen in a manner similar to HLA mismatches, where differences in antigen presentation or processing increase risk of TRM. Indeed, we found rs200092801 could induce strong binding of mutant TEX38 antigen peptides with MHC class I molecules, which possibly triggered downstream immune response to the mutant antigens and led to TRM. Furthermore, we also observed higher TRM frequency in the rs200092801-mismatched recipient-donor pairs when the pairs carried HLA class I alleles that could strongly bind with the mutant TEX38 antigen. Our results support that the increased TRM risk of allele mismatches at rs200092801 could be a result of differences in antigen presentation induced by the variant. Another important question is how this testis antigen is recognized by the immune system. It is likely that the TEX38 antigen is an NS-MGCA that can pass through the Sertoli cell barrier and be recognized by MHC class I molecules in immune cells. The TEX38 antigen may also be expressed in other cells, even cancer cells. Recently, a large number of proteins normally restricted to the germ cells of the testes were discovered to be expressed in various cancer cells, including melanoma, liver cancer, lung cancer, bladder cancer, and leukemia cells.29-31 These cancer/testis antigens have become promising targets in cancer immunotherapy.32-35 Whether the TEX38 antigen is expressed in recipients’ cancer cells and whether the latter also play a role in TRM in recipients are questions that remain to be explored. Of course, our novel findings regarding mismatches at a rare TEX38 variant increasing TRM risk need further replication, and if replicated, additional experimental work would be necessary to determine how this variant affects TRM.
The only recipient gene we found to be associated with OS, OR51D1, was driven by its association with TRM. Recipients carrying 1 of the 6 mutations in OR51D1 were protected from TRM. OR51D1 belongs to class I ORs, which may be specialized in detecting water-soluble odorants. Recently, OR51D1 and many other class I ORs were found to be expressed in blood leukocytes, and it was suggested that these receptors may equip the immune system to react to chemicals entering the bloodstream.36 In addition, ORs are expressed in the white blood cells of patients with AML and may be novel therapeutic targets for AML because OR activation leads to reduced cancer cell proliferation and/or induced apoptosis and erythrocyte differentiation.37,38 The effect of ORs on suppressing cancer cell growth has also been documented in liver cancer,39 prostate cancer,40,41 and non–small-cell lung cancer.42 Although the mechanism for how OR51D1 affect TRM after BMT is not known, this novel finding identifies a new direction for potentially reducing TRM and ultimately improving patient survival.
The finding of NT5E associated with DRM in donor genotypes is of interest because NT5E is a key metabolic regulator of several drugs used for BMT, including Flu and MMF. We first speculated that the effect of NT5E on DRM could be exerted through drug metabolism. However, we failed to determine whether taking the drugs (Flu or MMF) or not resulted in any difference in the effect of NT5E on DRM, because only 28 patients received transplants from donors carrying NT5E mutations, and in all cases, the patients did not die as a result of DRM within the first year after transplantation. Therefore, even larger cohorts than DISCOVeRY-BMT will be needed to evaluate whether drug usage makes any difference on the impact of NT5E on DRM. Alternatively, NT5E might affect DRM through its catalysis of adenosine monophosphate to adenosine, which is the major control point for extracellular adenosine levels. Numerous studies have revealed the immunosuppressive and proangiogenic effects of extracellular adenosine and the important role it plays in tumor onset and progression.22-26 NT5E overexpression has been found in various cancer types22,24 and is associated with poor prognosis.43-46 We found the rare NT5E mutations present in donors could have an impact on various aspects of enzyme activity, including protein conformation change, dimerization, and binding with adenosine (Figure 3). Therefore, we hypothesized that donors carrying those mutations had lower adenosine-generating efficiency, and hence, the recipients whose donors carried the mutations experienced NT5E inhibition. Recent preclinical studies have demonstrated that targeted blockade of NT5E can effectively inhibit tumor growth.46-55 Consistent with the reported therapeutic effect of NT5E suppression, we observed no disease relapse in our patients with blood cancer who received transplants from donors carrying NT5E rare mutations. This observation may provide clinical evidence supporting anti-NT5E therapy for patients with cancer. Future functional studies of these rare coding mutations and additional confirmation of our findings are needed to demonstrate this point.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) (R01 HL102278; principal investigators, T.H. and L.E.S.-C.). This work was also supported by National Cancer Institute (NCI) grant P30CA016056, involving the use of the Roswell Park Cancer Institute Bioinformatics and Biostatistics shared resources. The Center for International Blood and Marrow Transplant Research is supported by public health service grant/cooperative agreement 5U24-CA076518 from the NCI, NHLBI, and the National Institute of Allergy and Infectious Diseases; grant/cooperative agreement 5U10HL069294 from the NHLBI and NCI; contract HHSH250201200016C with the Health Resources and Services Administration (HRSA)/Department of Health and Human Services; 2 grants (N00014-15-1-0848 and N00014-16-1-2020) from the Office of Naval Research; and support from Alexion, Amgen, Inc.*; an anonymous donation to the Medical College of Wisconsin; Astellas Pharma US; AstraZeneca; Be the Match Foundation; Bluebird Bio, Inc.*; Bristol-Myers Squibb Oncology*; Celgene Corporation*; Cellular Dynamics International, Inc.; Chimerix, Inc.*; Fred Hutchinson Cancer Research Center; Gamida Cell Ltd.; Genentech, Inc.; Genzyme Corporation; Gilead Sciences, Inc.*; Health Research, Inc.; Roswell Park Cancer Institute; HistoGenetics, Inc.; Incyte Corporation; Janssen Scientific Affairs, LLC; Jazz Pharmaceuticals, Inc.*; Jeff Gordon Children’s Foundation; The Leukemia & Lymphoma Society; Medac, GmbH; MedImmune; The Medical College of Wisconsin; Merck & Co, Inc.*; Mesoblast; MesoScale Diagnostics, Inc.; Miltenyi Biotec, Inc.*; National Marrow Donor Program; Neovii Biotech NA, Inc.; Novartis Pharmaceuticals Corporation; Onyx Pharmaceuticals; Optum Healthcare Solutions, Inc.; Otsuka America Pharmaceutical, Inc.; Otsuka Pharmaceutical Co, Ltd. Japan; Patient-Centered Outcomes Research Institute; Perkin Elmer, Inc.; Pfizer, Inc; Sanofi US*; Seattle Genetics*; Spectrum Pharmaceuticals, Inc.*; St. Baldrick’s Foundation; Sunesis Pharmaceuticals, Inc.*; Swedish Orphan Biovitrum, Inc.; Takeda Oncology; Telomere Diagnostics, Inc.; University of Minnesota; and Wellpoint, Inc.* *Corporate members.
The views expressed in this article do not reflect the official policy or position of the NIH, the Department of the Navy, the Department of Defense, HRSA, or any other agency of the US Government.
Authorship
Contribution: Q.Z., L.Y., T.H. and L.E.S.-C. conceived the study and performed data analysis; Q.Z., T.H., and L.E.S.-C. wrote the manuscript; C.Z. and L.W. performed the crystal structure analysis and the major histocompatibility complex binding prediction, respectively; D.O.S., L. Pooler, X.S., and C.A.H. performed genotyping and assessed and interpreted genotype data; L. Preus, X.Z., S.R.S., M.P., T.H., and L.E.S.-C. acquired and organized clinical data; Q.Z., L.Y., Q.L., Q.H., L. Preus, and S.L. provided quality control and data randomization; Q.Z., L.Y., C.Z, A.I.C.-G., K.O., P.L.M, S.L., T.H., and L.E.S.-C. interpreted data; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qianqian Zhu, Department of Biostatistics & Bioinformatics, Roswell Park Cancer Institute, Elm & Carlton Streets, Buffalo, NY 14263; e-mail: qianqian.zhu@roswellpark.org; and Lara E. Sucheston-Campbell, College of Pharmacy, College of Veterinary Medicine, The Ohio State University, Columbus, OH 43210; e-mail: sucheston-campbell.1@osu.edu.
References
Author notes
Q.Z., L.Y., T.H., and L.E.S.-C. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal