In this issue of Blood, Zhu et al identify a novel rare genotype mismatch in the testis-expressed gene TEX38 that is prognostic of blood and marrow transplant (BMT) mortality (see figure).1 The single-nucleotide polymorphism (SNP) that was identified (rs200092801) in this first of its kind exome-wide association study (EXWAS) is a nonsynonymous variant, suggesting a functional consequence underlying this association. The hunt for prognostic and predictive biomarkers is a key component to precision medicine efforts, and the assessment by Zhu et al is an important step in that direction for those who receive BMT.
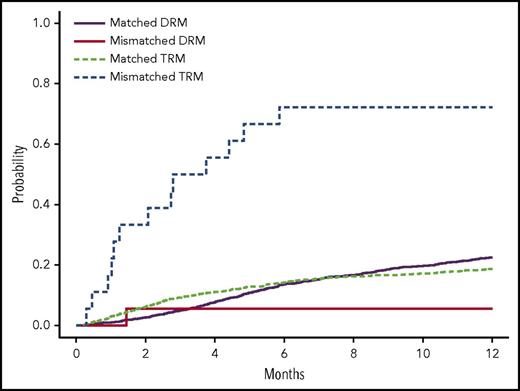
Estimated cumulative incidence curves with TRM and DRM as competing events for recipients with donor genotypes having 1 allele mismatch and no mismatch of rs200092801 in cohort 1. Two allele mismatches of this variant between recipients and donors were not observed. See Figure 1 in the article by Zhu et al that begins on page 2490.
Estimated cumulative incidence curves with TRM and DRM as competing events for recipients with donor genotypes having 1 allele mismatch and no mismatch of rs200092801 in cohort 1. Two allele mismatches of this variant between recipients and donors were not observed. See Figure 1 in the article by Zhu et al that begins on page 2490.
Notably, BMT remains a primary treatment option for those with malignant hematologic diseases, including acute lymphoblastic leukemia, acute myeloid leukemia, and myelodysplastic syndrome. Because of better donor selection, supportive care, and infection control, outcomes for individuals receiving BMT have improved in recent years. However, 1-year mortality is still ∼40%.2 An important predictor of outcome in BMT is matching patients with unrelated donors based on 4 human leukocyte antigen (HLA) genetic loci: HLA-A, HLA-B, HLA-C, and HLA-DRB1.2 In spite of the predictive power of these HLA genetic loci, there still remains significant interindividual variation in survival after BMT, suggesting there is more to learn in terms of predicting BMT outcome. Therefore, several studies have explored the role of non-HLA genetic loci on BMT survival.
There are multiple strategies available when identifying genetic variants associated with outcomes, and the “best” strategy depends on the research question. Zhu et al employed an EXWAS to identify variants and genes associated with overall survival (OS), transplant-related mortality (TRM), and disease-related mortality (DRM). In this scenario, a genotyping array (often called a chip) was used rather than whole-exome sequencing (WES). While WES provides greater coverage of the exome, the cost is still higher than using a genotyping array. This is particularly important when evaluating outcomes in the >4000 individuals included in the DISCOVeRY-BMT (Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT) study.3 The number of SNPs on the particular chip used in this analysis numbered >200 000. When conducting an EXWAS, the appropriate research question would be “what is the role of rare (eg, minor allele frequency [MAF] <0.5%) coding variants on a particular outcome?”4 This is in contrast to a genome-wide association study, where one would be evaluating the role of common genetic variants (eg, MAF >5%) across the entire genome (regardless of coding status), or a whole-genome sequencing study, where one typically evaluates the role of rare variants across the entire genome.4,5 Another approach would be a so-called candidate gene study. However, as the authors noted, candidate gene studies of BMT outcome have yield mixed results,6 which is true for multiple outcomes.5
This study contributes importantly to our understanding of the role of non-HLA genetic variants on BMT outcomes. As noted, the authors leveraged the DISCOVeRY-BMT study to conduct the first reported EXWAS of outcomes in individuals treated with BMT. An important methodologic aspect of this particular assessment was the evaluation of genetic variants in 2 independent cohorts: cohort 1 included 1970 recipients and 1741 donors, and cohort 2 included 503 recipients and 480 donors. Specifically, the authors found that donor–recipient mismatches for TEX38 rs200092801 were significantly (P = 3.51 × 10−7) associated with TRM. The effect was stronger when either the donor or recipient was female. Specifically, the median survival time for donor–recipient pairs mismatched at TEX38 rs200092801 was 1.1 months for female donor to female recipient, 0.9 months for female donor to male recipient, and 4.4 months for male donor to female recipient. These are dismal outcomes for these individuals. Unfortunately, as TEX38 rs200092801 is rare variant (MAF 0.3%), the minor allele was not observed in cohort 2, which is one potential limitation of the current study. However, the authors presented additional functional evidence in support of this their finding.
Aside from the single-variant association analyses, the authors also conducted gene-level association analyses, which aggregate information on multiple SNPs within a gene to make gene-level inferences. While not as useful for identifying single prognostic biomarkers, these gene-level approaches could provide insights to the underlying biology of poor outcomes among those who receive BMT. Genes identified in these analyses that were associated with OS, TRM, or DRM include OR51D1 (recipient), ALPP (donor), EMID1 (donor), SLC44A5 (donor), LRP1 (donor), HHAT (donor), LYZL4 (donor), and NT5E (donor).
What is next on the horizon for predicting poor outcomes for those who receive BMT? Of course there are several directions to move from the study by Zhu et al, but 4 important strategies would be: (1) Replicating the TEX38 rs200092801 finding in an independent population. This is particularly important as the effect of this variant could not be evaluated in cohort 2 of the present study. (2) Functionally validating TEX38 rs200092801. While the authors did provide some biological plausibility for their finding, much work is needed to understand why this variant is associated with TRM. (3) Applying genome-wide approaches for identifying variants that are prognostic of OS, TRM, and DRM. Common variants and/or variants in noncoding regions have been identified in relation to several treatment-related outcomes and diseases.5,7 It is important to assess the role of these variants to identify novel prognostic biomarkers of outcome in populations receiving BMT. (4) Evaluating genetic variants and BMT outcomes in non-European populations. While it is important to account for population stratification bias in genetic association studies,5 it is also vital to evaluate the role of these and other variants in other populations, which often have different responses to treatment.8 Ultimately, it is clear from this study that evaluating non-HLA genetic mismatches is an important strategy for predicting outcomes among those who receive BMT.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal