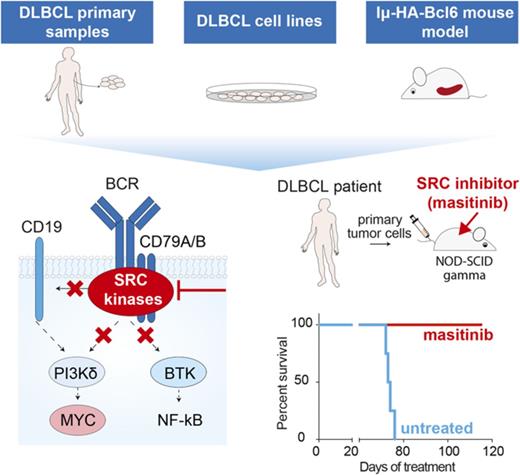
Key Points
Inhibition of BTK in patients who are resistant to ibrutinib changes signaling tumor dependencies and promotes MYC upregulation.
Multitarget inhibition of LYN, FYN, and BLK is therapeutically effective in patients with DLBCL independent of their molecular subtypes.
Abstract
In diffuse large B-cell lymphoma (DLBCL), activation of the B-cell receptor (BCR) promotes multiple oncogenic signals, which are essential for tumor proliferation. Inhibition of the Bruton’s tyrosine kinase (BTK), a BCR downstream target, is therapeutically effective only in a subgroup of patients with DLBCL. Here, we used lymphoma cells isolated from patients with DLBCL to measure the effects of targeted therapies on BCR signaling and to anticipate response. In lymphomas resistant to BTK inhibition, we show that blocking BTK activity enhanced tumor dependencies from alternative oncogenic signals downstream of the BCR, converging on MYC upregulation. To completely ablate the activity of the BCR, we genetically and pharmacologically repressed the activity of the SRC kinases LYN, FYN, and BLK, which are responsible for the propagation of the BCR signal. Inhibition of these kinases strongly reduced tumor growth in xenografts and cell lines derived from patients with DLBCL independent of their molecular subtype, advancing the possibility to be relevant therapeutic targets in broad and diverse groups of DLBCL patients.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive form of non-Hodgkin lymphoma. The molecular profile of patients diagnosed with DLBCL has unveiled intrinsic tumor differences hidden by the extremely similar histological appearance of the malignant tissues.1-3 In particular, transcriptional differences between DLBCL tumors led to the definition of 2 main subtypes: germinal center B-cell-like (GCB) and activated B-cell-like (ABC).1 The spectrum of prevalent mutations in these 2 subtypes reflects the different phases of B-cell maturation at the origin of these tumors.1,4-6 ABC DLBCL tumors are more prone to select mutations in genes regulating plasma cell differentiation and promoting the activity of NF-κB signaling.4,7-10 GCB DLBCLs typically show ectopic expression of the anti-apoptotic protein BCL2, as well as mutations of epigenetic modifiers, and several chromosomal alterations.4,11,12 This genetic diversity translates into different levels of tumor aggressiveness and response to therapies.13
Patients diagnosed with DLBCL, independent of their subtype, are primarily treated with combinations of an anti-CD20 antibody (rituximab) and generic chemotherapies, as the repertoire of targeted therapies available for this disease is still limited.13 Aberrant activation of B-cell receptor (BCR) signaling is one of the driver oncogenic events promoting B-cell proliferation in non-Hodgkin lymphoma.14 Stimulation of the BCR promotes the activation of multiple downstream targets, including BTK,15 the BCR co-receptor CD19,16 and PI3KCA/AKT.17 Recently, several inhibitors that block BCR oncogenic signals at different levels have been or are being tested in clinical trials.18-21 Notably, the therapeutic efficacy of these inhibitors varies between different types of non-Hodgkin lymphoma based on the cell of origin of the tumor and their dependencies on specific pathways downstream of the BCR. For example, clinical trials have shown that patients with DLBCL treated with ibrutinib, mainly an inhibitor of BTK,19 have a nonuniform response: patients classified as ABC subtype are frequently sensitive to BTK inhibition, whereas cases with a GCB molecular profile tend to not respond to the treatment.22 Although both ABC and GCB lymphoma depend on the activity of BCR,23,24 mutations in genes downstream of the BCR (eg, CD79 and MYD88) and genomic alterations, including mutations and chromosomal changes in genes involved in NF-κB signaling, are enriched preferentially in ABC DLBCL.8,9 Alterations in these genes facilitate chronic activation of BCR signaling,10 whereas the GCB subtype depends on the tonic activation of the BCR.23
In this study, we investigated whether BTK inhibition in ibrutinib-resistant tumors induces signal changes that may contribute to the lack of a therapeutic response. To this end, we explored whether blocking the propagation of the BCR oncogenic signal at its root could represent an effective therapeutic strategy for patients with DLBCL independent of their subtype and dependencies on specific signals.
Methods
Primary samples and cell culture
DLBCL primary samples were obtained from Dana-Farber Cancer Institute (the Public Repository of Xenografts) as cryopreserved vials after 1 passage in mice. For signaling assays, cells were plated at a concentration of 0.5 × 106 cells/mL in 10% fetal bovine serum RPMI, with dimethyl sulfoxide (DMSO) or 0.5 μM ibrutinib or 5 μM masitinib. Cells were harvested after 6 hours of treatment for the analysis of downstream signals. All cell lines were authenticated by STR DNA profiling. All cells were maintained in RPMI 1640 with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. Isolated mouse tumor B cells were maintained in culture for maximum 4 days in B-cell media (45% Dulbecco’s modified Eagle medium, 45% Iscove modified Dulbecco medium, 4 mM L-glutamine, 1% penicillin/streptomycin, 10% fetal bovine serum). See supplemental Methods (available on the Blood Web site) for more information.
In vivo patient-derived xenograft treatment studies
For in vivo studies, cells from the double-hit (DH) patient 69487 were infected with highly concentrated lentiviral particles (50-100 multiplicity of infection) and immediately IV inoculated in 6- to 8-week-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice. Transduced cells expressed luciferase and the mCherry-fluorescent marker. After the first engraftment, fluorescent positive cells were sorted and used for the following experiments. Cells from ABC DLBCL patient 13796 were directly IV inoculated in 6- to 8-week-old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice. For in vivo treatments using both patient-derived xenograft models, tumor cells were counted and 106 cells were IV xenografted into five 6- to 8-week-old NSG mice. One week after injection, mice were enrolled for treatment by daily intraperitoneal injection with ibrutinib (Chemietek, cat#CT-PCI327) diluted in 4% DMSO methylcellulose at 12 mg/kg per day, or 50 mg/kg masitinib (Abmole, cat#M1838) dissolved in 4% DMSO phosphate-buffered saline. Tumor growth was monitored by IVIS imaging or by ultrasound.
Results
BCR signaling in primary DLBCL patient samples before and after BTK inhibition
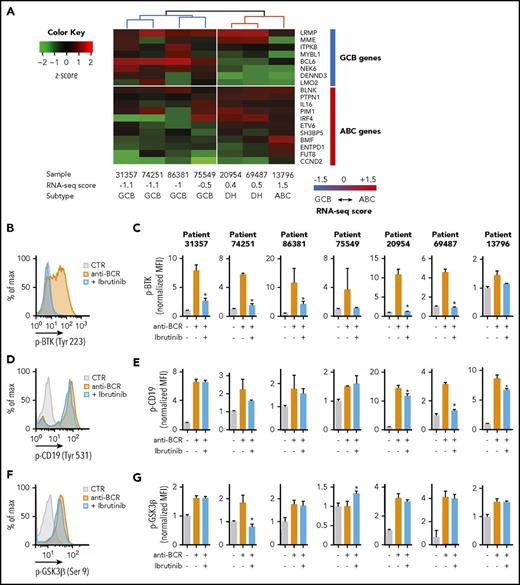
Pharmacologic inhibition of BTK results in the perturbation of multiple pathways, and its therapeutic efficacy in ABC-DLBCL depends on the inactivation of NF-κB signaling.25 However, it is not clear whether signaling changes also occur in patients who do not respond to ibrutinib treatment. We analyzed the activation of BCR signaling and the effect of BTK pharmacological inhibition in 7 DLBCL primary human samples.26 Purified lymphoma B cells were obtained from 3 patients who were newly diagnosed and had not yet received treatments and 4 patients who relapsed or were refractory to chemotherapies (supplemental Table 1). Cells were engrafted in NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) animals. Histological analyses of the engrafted tumors showed heterogeneous expression of CD10, BCL6, and MUM1 among the samples (supplemental Figure 1A). To categorize these samples into cell-of-origin subtypes, we analyzed their mRNA expression, as well as immunohistochemical and genetic features. Initially, we clustered the samples on the basis of the expression of 8 GCB-specific and 11 ABC-specific genes.6 Samples were assigned to each subtype on the basis of a computed RNA-seq score.6 In our cohort, 4 samples were classified as GCB-DLBCL and 3 samples as ABC-DLBCL (Figure 1A). However, we noticed that 2 samples (20954 and 69487) that were classified as ABC on the basis of their expression profile were actually DH (MYC-BCL2) and CD10-positive lymphomas; therefore, because of this discrepancy between expression and immunohistochemical and genetic features, we defined them as DH. The expression score for the other samples matched with their histopathological and genetic profiles. Targeted sequencing of frequently mutated genes in DLBCL (supplemental Table 2) confirmed the heterogeneous repertoire of genomic alterations typical of DLBCL (supplemental Table 3). Samples classified as GCB have mutations in MLL2, MLL3, EP300, or CREBBP, and rare or no alterations in genes involved in NF-κB signal (supplemental Table 3). In all patients, independent of their molecular profile, the basal phosphorylation levels of BTK (Tyr223) were similar to the unstained control in unstimulated condition (supplemental Figure 1B). Notably, activation of BCR using anti-BCR antibodies promoted the phosphorylation of BTK, CD19, and GSK3β in all tumors, except for phospho-GSK3β in sample 75549 (Figure 1B-G). Similarly, pharmacological inhibition of BTK with ibrutinib effectively blocked BTK phosphorylation (Figure 1B-C), but in the majority of the samples did not change CD19 (Figure 1D-E) and GSK3β phosphorylation (Figure 1F-G). Hence, activation of BCR induces multiple oncogenic signals, and treatment with ibrutinib prevents preferentially the activation of BTK in both patients with ABC-like and GCB-like molecular features.
Analysis of BCR signaling in primary DLBCL patient samples. (A) Classification of 7 DLBCL primary samples based on their expression profile. The heat map shows the expression (z-score) of 8 GCB and 11 ABC genes in the indicated samples. The computed RNA-seq score distinguishes between GCB, ABC DLBCL, or DH. (B,D,F) Representative flow cytometry analysis to determine the phosphorylation of BTK (B, patient 69487), CD19 (D, patient 20954), and GSK3β (F, patient 20954) in purified lymphoma cells stimulated with anti-BCR (orange) or treated with ibrutinib (blue). Unstimulated cells were used as control (gray). (C,E,G) Quantification of the median fluorescence values (MFIs) of pBTK (Tyr223) (C), pCD19 (Tyr 531) (E), and pGSK3β (Ser9) (G) in the indicated patients normalized to the stained, unstimulated control. Data are represented as bar plots corresponding to the mean ± standard deviation (SD) of 3 replicates. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars) are labeled with an asterisk (uncorrected P ≤ .05). Unlabeled bars indicate not statistically significant changes.
Analysis of BCR signaling in primary DLBCL patient samples. (A) Classification of 7 DLBCL primary samples based on their expression profile. The heat map shows the expression (z-score) of 8 GCB and 11 ABC genes in the indicated samples. The computed RNA-seq score distinguishes between GCB, ABC DLBCL, or DH. (B,D,F) Representative flow cytometry analysis to determine the phosphorylation of BTK (B, patient 69487), CD19 (D, patient 20954), and GSK3β (F, patient 20954) in purified lymphoma cells stimulated with anti-BCR (orange) or treated with ibrutinib (blue). Unstimulated cells were used as control (gray). (C,E,G) Quantification of the median fluorescence values (MFIs) of pBTK (Tyr223) (C), pCD19 (Tyr 531) (E), and pGSK3β (Ser9) (G) in the indicated patients normalized to the stained, unstimulated control. Data are represented as bar plots corresponding to the mean ± standard deviation (SD) of 3 replicates. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars) are labeled with an asterisk (uncorrected P ≤ .05). Unlabeled bars indicate not statistically significant changes.
To directly link BCR signaling with response or resistance to ibrutinib, we used a panel of well-characterized GCB and ABC lymphoma cell lines. Treatment with ibrutinib significantly decreased the cell survival of ABC-DLBCL cell lines (U2932 and HBL-1) and follicular lymphoma-transformed cell lines (DOHH2) (Figure 2A), but did not affect the cell viability of GCB cell lines (SU-DHL-6, VAL, OCI-LY19, and SU-DHL-10) and 1 ABC cell line (SU-DHL-2) harboring a mutation in MYD88 and biallelic A20 inactivation7 but CD79A/B wild-type8,22 (Figure 2B). As it was observed in the primary patient samples, phospho-BTK was almost undetectable in unstimulated cells independent of their subtype or whether they harbored mutations in CD79, MYD88, or other genes involved in NF-κB activation8-10,22,27,28 (supplemental Figure 2A). In cell lines stimulated with antibodies to mimic the activation of the BCR, treatment with ibrutinib could equally block BTK phosphorylation in GCB and ABC lymphomas (Figure 2C; supplemental Figure 2B). However, it did not decrease the phosphorylation of CD19 (Figure 2D-E; supplemental Figure 2C) and GSK3β, which remained active in both GCB and ABC cells treated with ibrutinib (Figure 2F-G; supplemental Figure 2D). Thus, activation of BCR signaling and inhibition of BTK lead to similar signaling changes in both ABC and GCB DLBCL, indicating that the opposite response to the treatment is associated with different downstream effectors.
Treatment with ibrutinib changes MYC expression in cells sensitive and resistant to ibrutinib. (A-B) Percentage of viable cells in the indicated lymphoma cell lines treated with DMSO or 0.1 or 0.5 μM ibrutinib for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a bar graph corresponding to the mean ± SD. (C) Western blot analysis of pBTK levels in GCB and ABC cells after DMSO or ibrutinib treatment (0.5 μM, 6 hours) and BCR stimulation (20 μg/mL F[ab']2 anti-human immunoglobulin M, 3 min). (D-G) Phosphoflow cytometry analysis and quantification of CD19 (Tyr 531) (D-E) and GSK3β (Ser 9) (F-G) in stimulated lymphoma cells pretreated with DMSO or ibrutinib (0.5 μM) for 6 hours, normalized to the stained, unstimulated controls. (H) MYC expression in the indicated ibrutinib-resistant cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates. Data have been normalized to the DMSO-treated control and are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib-treated cells were labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. (I) Western blot analysis of the indicated ibrutinib-resistant cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. (J) MYC expression in the indicated ibrutinib-sensitive cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates. Data have been normalized to the DMSO-treated control and are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib-treated cells were labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. (K) Western blot analysis of the indicated ibrutinib-sensitive cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. (L) Western blot analysis of MYC and BTK in isogenic GCB cell lines WT or BTK KO. (M) Heat map indicating the expression levels (z-scores) of 10 MYC target genes in WSU-DLCL2 cells WT or BTK KO.
Treatment with ibrutinib changes MYC expression in cells sensitive and resistant to ibrutinib. (A-B) Percentage of viable cells in the indicated lymphoma cell lines treated with DMSO or 0.1 or 0.5 μM ibrutinib for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a bar graph corresponding to the mean ± SD. (C) Western blot analysis of pBTK levels in GCB and ABC cells after DMSO or ibrutinib treatment (0.5 μM, 6 hours) and BCR stimulation (20 μg/mL F[ab']2 anti-human immunoglobulin M, 3 min). (D-G) Phosphoflow cytometry analysis and quantification of CD19 (Tyr 531) (D-E) and GSK3β (Ser 9) (F-G) in stimulated lymphoma cells pretreated with DMSO or ibrutinib (0.5 μM) for 6 hours, normalized to the stained, unstimulated controls. (H) MYC expression in the indicated ibrutinib-resistant cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates. Data have been normalized to the DMSO-treated control and are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib-treated cells were labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. (I) Western blot analysis of the indicated ibrutinib-resistant cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. (J) MYC expression in the indicated ibrutinib-sensitive cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates. Data have been normalized to the DMSO-treated control and are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib-treated cells were labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. (K) Western blot analysis of the indicated ibrutinib-sensitive cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. (L) Western blot analysis of MYC and BTK in isogenic GCB cell lines WT or BTK KO. (M) Heat map indicating the expression levels (z-scores) of 10 MYC target genes in WSU-DLCL2 cells WT or BTK KO.
MYC expression correlates with DLCBL resistance to ibrutinib
MYC is a common downstream target of IRF4,25,29,30 CD19,31,32 and GSK3β.33-35 Surprisingly, we observed that sensitivity and resistance to ibrutinib in ABC and GCB lymphomas was associated with changes in MYC expression, independent of MYC basal levels in the cells (supplemental Figure 2E). Indeed, GCB and ABC cells (SU-DHL-2) resistant to ibrutinib exhibited a significant upregulation of MYC upon treatment (Figure 2H-I). Conversely, cells sensitive to BTK inhibition downregulated both MYC and IRF4 (Figure 2J-K). To link BTK inactivation with MYC expression and exclude ibrutinib off-target effects, we genetically knocked-out BTK in ABC and GCB cells using an inducible Cas9 and CRISPRs expressed in tandem with a fluorescent marker. ABC cells (U2932 and HBL-1) were highly sensitive to BTK disruption, and fluorescent-positive cells rapidly disappeared after doxycycline-induced Cas9 expression (supplemental Figure 2F). Conversely, GCB-derived cells (WSU-DLCL2 and HT) tolerated the absence of BTK, and we could clonally expand and select BTK null cells (supplemental Figure 2G-H). In 2 independent clones for each cell line, we observed that a complete knockout of BTK was complemented by MYC overexpression (Figure 2L). Moreover, BTK-null cells proliferated more than the parental lines (supplemental Figure 2I), and they significantly upregulated the expression of genes involved in cell proliferation and DNA replication (supplemental Figure 2J). Importantly, several upregulated genes promoting cell cycle progression (eg, E2F1 and CDC25A) were previously reported MYC direct targets36-39 (Figure 2M). Hence, inhibition of BTK is therapeutically effective in BTK-dependent lymphomas, but leads to the upregulation of MYC oncogenic signaling in BTK-independent tumors.
MYC is upregulated in Iμ-HA-Bcl6 animals treated with ibrutinib
To explore whether BTK inhibition causes a similar response in vivo, we tested the effect of ibrutinib in the Iμ-HA-Bcl6 mouse model of lymphoma that has been used to study DLBCL pathogenesis.40-42 Approximately 75% to 80% of Iμ-HA-Bcl6 transgenic animals are reported to develop lymphoproliferative disease or lymphoma associated with splenomegaly within 15 to 20 months as a result of constitutive Bcl6 overexpression. We monitored tumor development in Iμ-HA-Bcl6 animals by measuring the spleen size, using ultrasound. Most of the animals between 13 and 21 months showed abnormal enlargement of the spleen that varied between 15 and 22 mm, whereas wild-type animals of the same age had a spleen size of 13 mm (supplemental Figure 3A). Histologic analysis confirmed the presence of large cells and expansion of the germinal centers enriched in B220-positive cells, together with heterogeneous detection of Bcl6 and PNA (supplemental Figure 3B). Characterization of splenic cells revealed enrichment of CD23-negative, immunoglobulin M-high/immunoglobulin D-low B cells in transgenic animals compared with the wild-type (supplemental Figure 3C).
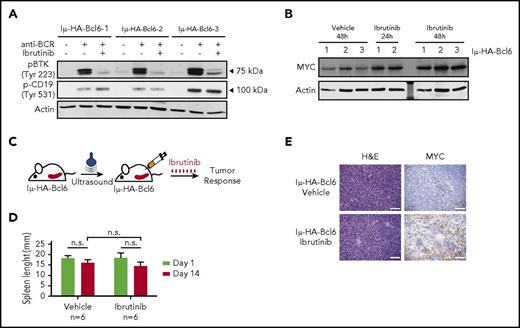
Next, we isolated Iμ-HA-Bcl6 B220+CD19+ lymphoma cells and stimulated them with anti-BCR. Purified B cells concurrently activated BCR downstream signals, inducing both BTK and CD19 phosphorylation (Figure 3A). Treatment with ibrutinib effectively blocked BTK phosphorylation (Figure 3A), but did not impair cell viability (supplemental Figure 3D), and CD19 signaling remained active (Figure 3A), indicating that BTK is dispensable for lymphoma Iμ-HA-Bcl6 B cell survival. Importantly, as previously observed in human DLBCL cells not responsive to ibrutinib, murine lymphomas treated with ibrutinib for 24 and 48 hours upregulated Myc protein (Figure 3B).
MYC is upregulated in IμHABCL6 animals harboring ibrutinib-resistant DLBCL. (A) Western blot analysis of pBTK (Tyr223) and pCD19 (Tyr531) in purified murine B-cells pretreated with DMSO or ibrutinib (0.5 μM) for 6 hours and stimulated with H2O2 for 3 minutes. (B) Western blot analysis of MYC levels in purified tumor B cells from 18- to 20-month-old IμHABCL6 mice. Cells isolated from each animal were divided in 3 aliquots and kept in culture in the presence of DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. (C) Pipeline for ibrutinib treatment in IμHABCL6 mice. Spleen size was assessed by ultrasound in 13- to 21-month-old mice. Mice enrolled in the study were treated with ibrutinib (12 mg/kg per day IP) for 14 days. (D) Spleen length in IμHABCL6 mice measured by ultrasound at day 1 and 14 in animals treated with ibrutinib or vehicle. Data are shown as a bar graph corresponding to the mean of 6 mice ± SD. P values were calculated using 2-tailed Student t test. (E) Representative images of immunohistochemistry analyses of IμHABCL6 mice spleen tissues. Scale bars, 100 μm. H&E, hematoxylin and eosin; n.s., not statistically significant.
MYC is upregulated in IμHABCL6 animals harboring ibrutinib-resistant DLBCL. (A) Western blot analysis of pBTK (Tyr223) and pCD19 (Tyr531) in purified murine B-cells pretreated with DMSO or ibrutinib (0.5 μM) for 6 hours and stimulated with H2O2 for 3 minutes. (B) Western blot analysis of MYC levels in purified tumor B cells from 18- to 20-month-old IμHABCL6 mice. Cells isolated from each animal were divided in 3 aliquots and kept in culture in the presence of DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. (C) Pipeline for ibrutinib treatment in IμHABCL6 mice. Spleen size was assessed by ultrasound in 13- to 21-month-old mice. Mice enrolled in the study were treated with ibrutinib (12 mg/kg per day IP) for 14 days. (D) Spleen length in IμHABCL6 mice measured by ultrasound at day 1 and 14 in animals treated with ibrutinib or vehicle. Data are shown as a bar graph corresponding to the mean of 6 mice ± SD. P values were calculated using 2-tailed Student t test. (E) Representative images of immunohistochemistry analyses of IμHABCL6 mice spleen tissues. Scale bars, 100 μm. H&E, hematoxylin and eosin; n.s., not statistically significant.
Then, we investigated whether systemic treatment with ibrutinib can lead to similar accumulation of Myc in vivo. Iμ-HA-Bcl6 animals with a spleen larger than 15 mm were treated with ibrutinib (n = 6, 12 mg/kg intraperitoneally [IP]) or vehicle (n = 6) for 2 weeks (Figure 3C). At the end point of the trial, macroscopic inspection of the spleen did not denote differences in organ size (Figure 3D), and histopathological analyses of the tissues showed no changes in the tissue composition and apoptosis in treated and untreated animals (supplemental Figure 3E-F). Interestingly, we observed Myc upregulation in the spleen of animals treated with ibrutinib (Figure 3E) coupled with an increase in Myc expression in purified B cells (supplemental Figure 3G). Thus, we confirmed that inhibition of BTK induced MYC upregulation in murine B-cell lymphoma resistant to ibrutinib.
Dual inhibition of BTK and PI3Kδ prevents MYC upregulation
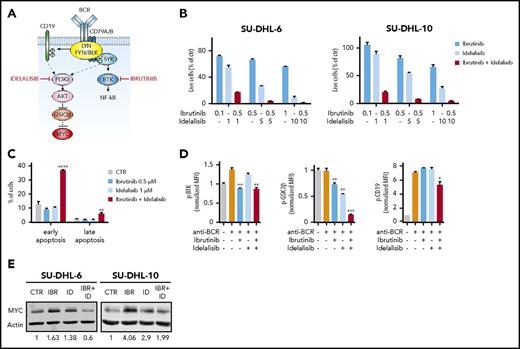
Thus far, we showed that activation of BCR signaling leads to the phosphorylation of multiple downstream proteins such as BTK, CD19, and GSK3β. Selective inhibition of BTK does not block other BCR targets and promotes MYC upregulation in tumors insensitive to BTK blockade. Thus, we wondered whether, by blocking additional signals downstream of the BCR, we could induce sensitivity to BTK inhibition and prevent MYC upregulation (Figure 4A). We combined ibrutinib with a specific PI3Kδ inhibitor (idelalisib) that has been recently approved for the treatment of specific lymphoma and leukemia subtypes.43-45 Interestingly, DLBCL cells (SU-DHL-10 and SU-DHL-6) with a poor response to ibrutinib or idelalisib as a single treatment were exquisitely sensitive to the drugs in combination (Figure 4B). Inhibition of BTK and PI3Kδ acted synergistically to promote apoptosis and to reduce cell proliferation at all tested doses (Figure 4B-C). As expected, treatment with idelalisib did not affect BTK and CD19 phosphorylation, but reduced phospho-GSK3β. In contrast, the combination of ibrutinib and idelalisib blocked both BTK and GSK3β activation and had a mild effect on phospho-CD19 (Figure 4D). Importantly, combined treatment with ibrutinib and idelalisib prevented MYC upregulation (Figure 4E). Hence, hampering multiple signals downstream of BCR is essential in BTK-independent tumors to promote the response to therapies and block the activation of MYC.
Synergism between BTK and PI3Kδ inhibition in ibrutinib-resistant GCB lymphoma cells. (A) Schematic representation BCR signaling. Ibrutinib and idelalisib block 2 effectors downstream of the BCR. (B) Percentage of viable cells in the indicated GCB lymphoma cell lines, treated with DMSO, ibrutinib, and idelalisib as single agents or in combination, at the indicated concentrations, for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a bar graph corresponding to the mean ± SD. (C) Induction of apoptosis in SU-DHL-10 cell lines treated with ibrutinib and idelalisib as single agents or in combination at the indicated concentrations. Data are shown as a bar graph corresponding to the mean ± SD of 3 replicates. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib- and/or idelalisib-treated cells were labeled with **P ≤ .01; ****P ≤ .0001. (D) Phosphoflow cytometry analysis and quantification of pBTK (Tyr223), pCD19 (Tyr 531), and pGSK3β (Ser 9) in stimulated SU-DHL-10 cells pretreated with DMSO or ibrutinib (0.5 μM) and/or idelalisib (1 μM) for 6 hours. Data are represented as bar plots corresponding to the mean ± SD of 3 replicates, normalized to the stained, unstimulated controls. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars), idelalisib (light blue bars), or a combination of the 2 (red bars) are labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. Unlabeled bars indicate not statistically significant changes. (E) Western blot analysis of the indicated cell lines treated with DMSO or ibrutinib (0.5 μM) and/or idelalisib (1 μM) for 24 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. CTR, control; IBR, ibrutinib; ID, idelalisib.
Synergism between BTK and PI3Kδ inhibition in ibrutinib-resistant GCB lymphoma cells. (A) Schematic representation BCR signaling. Ibrutinib and idelalisib block 2 effectors downstream of the BCR. (B) Percentage of viable cells in the indicated GCB lymphoma cell lines, treated with DMSO, ibrutinib, and idelalisib as single agents or in combination, at the indicated concentrations, for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a bar graph corresponding to the mean ± SD. (C) Induction of apoptosis in SU-DHL-10 cell lines treated with ibrutinib and idelalisib as single agents or in combination at the indicated concentrations. Data are shown as a bar graph corresponding to the mean ± SD of 3 replicates. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib- and/or idelalisib-treated cells were labeled with **P ≤ .01; ****P ≤ .0001. (D) Phosphoflow cytometry analysis and quantification of pBTK (Tyr223), pCD19 (Tyr 531), and pGSK3β (Ser 9) in stimulated SU-DHL-10 cells pretreated with DMSO or ibrutinib (0.5 μM) and/or idelalisib (1 μM) for 6 hours. Data are represented as bar plots corresponding to the mean ± SD of 3 replicates, normalized to the stained, unstimulated controls. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars), idelalisib (light blue bars), or a combination of the 2 (red bars) are labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. Unlabeled bars indicate not statistically significant changes. (E) Western blot analysis of the indicated cell lines treated with DMSO or ibrutinib (0.5 μM) and/or idelalisib (1 μM) for 24 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. CTR, control; IBR, ibrutinib; ID, idelalisib.
Genetic and pharmacological inhibition of SRC kinases acting upstream of BTK and PI3Kδ
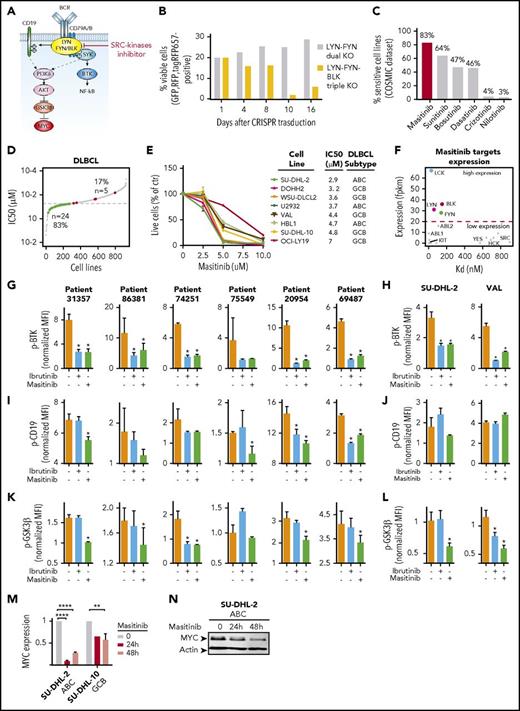
We then wondered whether targeting the SRC kinases responsible for propagating the BCR signal to its downstream effectors would be therapeutically effective in all DLBCLs, independent of their molecular subtypes and signaling dependencies. LYN is the first protein directly activated by the BCR,14 and it catalyzes the transmission of the signal to multiple effectors, including SYK, BTK, and CD19 (Figure 5A). However, other SRC family kinases, such as FYN and BLK, can replace LYN during B-cell differentiation.46,47 We observed a similar redundancy of these SRC kinases in DLBCL. Indeed, in SU-DHL-2 cells, neither single CRISPR/Cas9-mediated knock-out of LYN, FYN, and BLK (supplemental Figure 4A-C), nor the dual knockout of LYN and FYN, had an effect on cell proliferation (supplemental Figure 4D). Importantly, only the triple knockout of LYN, FYN, and BLK significantly reduced the number of viable cells (Figure 5B), indicating that it is necessary to block the activity of all these 3 kinases to hinder tumor growth.
Genetic and pharmacological inhibition of SRC-family kinases block multiple oncogenic signals downstream BCR. (A) BCR signaling representation. The SRC family kinases LYN, FYN, and BLK transmit the signal to multiple effectors including SYK-BTK, CD19, and GSK3β. (B) Quantification of cell viability in SU-DHL-2 cells with dual or triple knockout of LYN, FYN, and BLK, based on the percentage of fluorescent cells (GFP, RFP, tagRFP657). Dual LYN-FYN (RFP, GFP-positive) cells were used as control. (C) Percentage of cells sensitive to the indicated SRC inhibitors based on COSMIC dataset. (D) Scatter plot representing the IC50s (the dotted line indicates the 50% inhibitory concentration (IC50) threshold = 20 μM) for masitinib treatment in 923 cells lines (gray). The colored points represent the DLBCL cell lines. In green are the cell lines sensitive to masitinib (24 cell lines) with IC50 lower than the threshold, and in red are the cell lines resistant to masitinib (5 cell lines), with IC50 higher than the threshold. (E) Percentage of viable cells in the indicated ABC and GCB lymphoma cell lines, treated with DMSO or with masitinib at 2.5, 5, or 10 μM for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a graph corresponding to the mean ± SD. (F) Scatter plot showing the expression of masitinib target genes (fpkm, fragments per kilobase million) vs their reported dissociation constant (Kd) for masitinib. (G,I,K) Quantification of fluorescence signals (MFIs) of BTK (G), CD19 (I), and GSK3β (K) phosphorylation assessed by phosphoflow cytometry for patients with DLBCL treated with DMSO or ibrutinib (0.5 μM) or masitinib (5 μM) for 6 hours and stimulated with H2O2 for 3 minutes. The bar plots correspond to the mean of normalized MFI ± SD of 3 replicates, and data were normalized on stained, unstimulated cells. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars) or masitinib (green bars) are labeled with an asterisk (uncorrected P ≤ .05). Unlabeled bars indicate not statistically significant changes. (H,J,L) Quantification of BTK (H), CD19 (J), and GSK3β (L) phosphorylation fluorescence signals assessed by phospho-flow cytometry for indicated cell lines treated with DMSO or ibrutinib (0.5 μM) or masitinib (5 μM) for 6 hours and stimulated with H2O2 for 3 minutes. The bar plots correspond to the mean of normalized MFI ± SD of 3 replicates, normalized on stained, unstimulated controls. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars) or masitinib (green bars) are labeled with an asterisk (P ≤ .05). Unlabeled bars indicate not statistically significant changes. (M) MYC expression in the indicated ibrutinib-resistant cell lines treated with DMSO or masitinib (5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates, and data are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and masitinib-treated cells are labeled with **P ≤ .01; ****P ≤ .0001. (N) Western blot analysis of MYC in SU-DHL-2 cells ibrutinib-resistant treated with DMSO or masitinib (5 μM) for 24 or 48 hours.
Genetic and pharmacological inhibition of SRC-family kinases block multiple oncogenic signals downstream BCR. (A) BCR signaling representation. The SRC family kinases LYN, FYN, and BLK transmit the signal to multiple effectors including SYK-BTK, CD19, and GSK3β. (B) Quantification of cell viability in SU-DHL-2 cells with dual or triple knockout of LYN, FYN, and BLK, based on the percentage of fluorescent cells (GFP, RFP, tagRFP657). Dual LYN-FYN (RFP, GFP-positive) cells were used as control. (C) Percentage of cells sensitive to the indicated SRC inhibitors based on COSMIC dataset. (D) Scatter plot representing the IC50s (the dotted line indicates the 50% inhibitory concentration (IC50) threshold = 20 μM) for masitinib treatment in 923 cells lines (gray). The colored points represent the DLBCL cell lines. In green are the cell lines sensitive to masitinib (24 cell lines) with IC50 lower than the threshold, and in red are the cell lines resistant to masitinib (5 cell lines), with IC50 higher than the threshold. (E) Percentage of viable cells in the indicated ABC and GCB lymphoma cell lines, treated with DMSO or with masitinib at 2.5, 5, or 10 μM for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a graph corresponding to the mean ± SD. (F) Scatter plot showing the expression of masitinib target genes (fpkm, fragments per kilobase million) vs their reported dissociation constant (Kd) for masitinib. (G,I,K) Quantification of fluorescence signals (MFIs) of BTK (G), CD19 (I), and GSK3β (K) phosphorylation assessed by phosphoflow cytometry for patients with DLBCL treated with DMSO or ibrutinib (0.5 μM) or masitinib (5 μM) for 6 hours and stimulated with H2O2 for 3 minutes. The bar plots correspond to the mean of normalized MFI ± SD of 3 replicates, and data were normalized on stained, unstimulated cells. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars) or masitinib (green bars) are labeled with an asterisk (uncorrected P ≤ .05). Unlabeled bars indicate not statistically significant changes. (H,J,L) Quantification of BTK (H), CD19 (J), and GSK3β (L) phosphorylation fluorescence signals assessed by phospho-flow cytometry for indicated cell lines treated with DMSO or ibrutinib (0.5 μM) or masitinib (5 μM) for 6 hours and stimulated with H2O2 for 3 minutes. The bar plots correspond to the mean of normalized MFI ± SD of 3 replicates, normalized on stained, unstimulated controls. Significant changes between stimulated cells (orange bars) and stimulated cells treated with ibrutinib (blue bars) or masitinib (green bars) are labeled with an asterisk (P ≤ .05). Unlabeled bars indicate not statistically significant changes. (M) MYC expression in the indicated ibrutinib-resistant cell lines treated with DMSO or masitinib (5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates, and data are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and masitinib-treated cells are labeled with **P ≤ .01; ****P ≤ .0001. (N) Western blot analysis of MYC in SU-DHL-2 cells ibrutinib-resistant treated with DMSO or masitinib (5 μM) for 24 or 48 hours.
Thus, we performed an in silico screen to identify pharmacologic inhibitors that could simultaneously block the activity of multiple SRC kinases. First, we selected 16 SRC inhibitors that were either approved by the US Food and Drug Administration or were in clinical trials and that could bind to LYN, FYN, and BLK with a dissociation constant (Kd) <2 μM, as a proxy for drug specificity48 (supplemental Table 5). Then, we filtered these inhibitors for their therapeutic efficacy in DLBCL, using screening response data from the COSMIC Genomics of Drug Sensitivity in Cancer database.27 Interestingly, masitinib scored as the most effective compound targeting DLBCL (Figure 5C; supplemental Table 5). Briefly, of the 923 cell lines tested for the response to masitinib, 32% (296/923) were considered sensitive (supplemental Figure 4E). Strikingly, among DLBCL cell lines, 83% (24/29) of the cells were sensitive to the drug (P = 1.5E−8) (Figure 5D). Moreover, 64% of the DLBCL cells were sensitive to sunitinib, 47% to bosutinib, and 46% to dasatinib (Figure 5C; supplemental Figure 4F-G), indicating an exquisite sensitivity of DLBCL cells to SRC inhibitors, and masitinib in particular. We confirmed that treatment with masitinib, bosutinib, or dasatinib effectively reduced the viability of DLBCL cells independent of their molecular subtype or responsiveness to ibrutinib in vitro (Figure 5E; supplemental Figure 4H-I). To establish the target specificity of these inhibitors, we correlated the expression of the masitinib-, bosutinib-, and dasatinib-predicted targets with their Kd values. DLBCL cell lines (OCI-LY19, SU-DHL-10, and WSU-DLCL2) expressed only a subset of masitinib, bosutinib, and dasatinib targets. Interestingly, LYN, FYN, BLK, and LCK were the most highly expressed targets with a low Kd (Figure 5F; supplemental Figure 4J-K; supplemental Table 6), whereas, for example, KIT, which was previously reported as one of the main targets of masitinib,49 was very weakly expressed in these cells. Moreover, we noticed that masitinib at low Kd specifically binds LYN, FYN, BLK, and LCK, whereas bosutinib and dasatinib bind a broader spectrum of SRC-kinases with similarly low Kd, including SRC, YES, and HCK (Figure 5F; supplemental Figure 4J-K).
To demonstrate the selectivity of masitinib in targeting LYN, FYN, and BLK, we modeled their tridimensional structure and predicted the binding conformation of masitinib in the catalytic pocket of those kinases by protein-ligand docking. Masitinib appeared to bind LYN as a type II inhibitor, forming canonical hydrogen bonds with several specific residues in the hinge region and protruding below the activation loop (supplemental Figure 4L-N; supplemental Table 7).
Importantly, LYN shares 70% and 72% sequence identity with FYN and BLK, respectively, and they have highly similar tridimensional structures (supplemental Figure 4O). Considering only the residues in direct contact with masitinib, the sequence similarity increases to 96% (FYN) and 93% (BLK), and the different amino acids are either conserved (Leu62 of LYN is replaced by Val in BLK and Ile in FYN) or at the border of the binding site (Tyr132 of LYN is replaced by Ser in BLK) with minimum interactions with masitinib. Therefore, the calculated binding modes of masitinib with LYN, BLK, and FYN are identical (supplemental Figure 4P), suggesting that masitinib will simultaneously inhibit the activity of those 3 kinases with similar affinity and block BCR signaling initiation. Hence, we decided to use masitinib as a representative pan-SRC inhibitor for DLBCL.
Masitinib blocks multiple signals downstream of BCR
Next, we compared the ability of ibrutinib and masitinib to inhibit BCR downstream signaling in DLBCL primary samples and cell lines. Importantly, treatment with masitinib not only blocked the activation of BTK with similar efficacy of ibrutinib (Figure 5G-H; supplemental Figure 4Q) but also significantly reduced the phosphorylation of CD19 (Figure 5I-J) and GSK3β (Figure 5K-L) in most of the human primary samples and cell lines. Moreover, both ABC and GCB cells treated with masitinib significantly downregulated MYC (Figure 5M-N). Masitinib, by inhibiting the activity of multiple SRC kinases, hampers the transmission of the BCR oncogenic signal, and DLBCL cells are particularly susceptible to its pharmacologic activity.
Masitinib is therapeutically effective in DLBCL patient-derived xenograft models
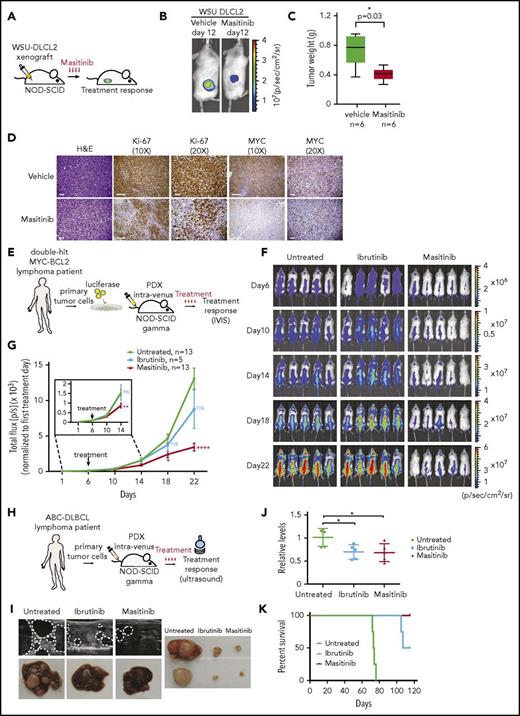
To explore the possibility of using masitinib as a novel therapeutic agent for the treatment of patients with DLBCL, we preclinically tested the efficacy of masitinib in cell lines and primary patient-derived xenograft models (Figure 6A, 6E and 6H). Initially, we subcutaneously inoculated WSU-DLCL2 cells expressing luciferase in NOD-SCID animals. Daily treatment with masitinib (n = 6 treated with vehicle and n = 6 treated with masitinib 50 mg/kg IP) significantly (P = .02) reduced tumor growth after 12 days of treatment (Figure 6B; supplemental Figure 5A). We observed a consistent reduction of the tumor mass in animals treated with masitinib (P = .03) (Figure 6C). Histopathological analysis of the residual tumor showed reduction of cell proliferation, as evidenced by ki-67 staining and downregulation of MYC (Figure 6D), but not a significant increase in the number of apoptotic cells (supplemental Figure 5B), indicating that masitinib reduces cell proliferation but it is insufficient to induce cell death.
Patient-derived cells engrafted in NSG mice are sensitive to masitinib. (A) Pipeline for xenograft studies. WSU-DLCL2 cells were injected subcutaneously; mice were treated with vehicle or masitinib (50 mg/kg per day IP) for 12 days, and tumor growth was assessed by luminescence. (B) Representative images of bioluminescence signals from xenografted mice after 12 days of treatment with vehicle or masitinib. (C) Average of the tumor weight harvest from animals treated with masitinib (n = 6) or vehicle (n = 6). The P value was calculated using 2-tailed Student t test. (D) Representative images of immunohistochemistry analyses of tumors harvested from xenografted animals and stained for Ki67 and MYC. Scale bars, 100 μm. (E) Representation of the experiment design to assess the effect of masitinib and ibrutinib in a DH patient-derived xenograft model (patient 69487). Primary tumor cells were obtained from a de novo diagnosed DH lymphoma patient, engineered to express luciferase and injected in NSG mice. Six days after injection, animals were treated with vehicle, masitinib (50 mg/kg per day IP), or ibrutinib (12 mg/kg per day IP), and tumor growth was assessed by luminescence. (F) Images of bioluminescent signals in animals treated with masitinib, ibrutinib, or vehicle at days 6, 10, 14, 18, and 22. (G) Quantification of the bioluminescence signals in animals treated with masitinib, ibrutinib, or vehicle at days 6, 10, 14, 18, and 22. The signal (photons/sec) was normalized to the first day of treatment (day 6). Tumor growth is represented as the mean ± SEM of the luminescence signal for the indicated number of animals. P value was calculated using 2-tailed Student t test. Significant changes between untreated and ibrutinib or masitinib-treated mice at the different points are labeled with *P ≤ .05; **P ≤ .01; ****P ≤ .0001. (H) Representation of the experimental design to assess the effect of masitinib and ibrutinib in ABC-DLBCL patient-derived xenograft (patient 13796). (I) Representative images by ultrasound of the abdomens and liver autopsy of animals harboring ABC lymphoma and treated vehicle, ibrutinib, and masitinib for 2 months. The dotted lines were designed to highlight the tumor. (J) MYC expression in blood samples isolated from animals untreated or treated with ibrutinib or masitinib (n = 5 per group). P values were calculated using 2-tailed Student t test. Significant changes between untreated and ibrutinib or masitinib-treated mice are labeled with *P ≤ .05. (K) Survival analysis of animals bearing ABC-lymphoma (patient 13796) untreated or treated with ibrutinib (12 mg/kg per day IP) or masitinib (50 mg/kg per day IP).
Patient-derived cells engrafted in NSG mice are sensitive to masitinib. (A) Pipeline for xenograft studies. WSU-DLCL2 cells were injected subcutaneously; mice were treated with vehicle or masitinib (50 mg/kg per day IP) for 12 days, and tumor growth was assessed by luminescence. (B) Representative images of bioluminescence signals from xenografted mice after 12 days of treatment with vehicle or masitinib. (C) Average of the tumor weight harvest from animals treated with masitinib (n = 6) or vehicle (n = 6). The P value was calculated using 2-tailed Student t test. (D) Representative images of immunohistochemistry analyses of tumors harvested from xenografted animals and stained for Ki67 and MYC. Scale bars, 100 μm. (E) Representation of the experiment design to assess the effect of masitinib and ibrutinib in a DH patient-derived xenograft model (patient 69487). Primary tumor cells were obtained from a de novo diagnosed DH lymphoma patient, engineered to express luciferase and injected in NSG mice. Six days after injection, animals were treated with vehicle, masitinib (50 mg/kg per day IP), or ibrutinib (12 mg/kg per day IP), and tumor growth was assessed by luminescence. (F) Images of bioluminescent signals in animals treated with masitinib, ibrutinib, or vehicle at days 6, 10, 14, 18, and 22. (G) Quantification of the bioluminescence signals in animals treated with masitinib, ibrutinib, or vehicle at days 6, 10, 14, 18, and 22. The signal (photons/sec) was normalized to the first day of treatment (day 6). Tumor growth is represented as the mean ± SEM of the luminescence signal for the indicated number of animals. P value was calculated using 2-tailed Student t test. Significant changes between untreated and ibrutinib or masitinib-treated mice at the different points are labeled with *P ≤ .05; **P ≤ .01; ****P ≤ .0001. (H) Representation of the experimental design to assess the effect of masitinib and ibrutinib in ABC-DLBCL patient-derived xenograft (patient 13796). (I) Representative images by ultrasound of the abdomens and liver autopsy of animals harboring ABC lymphoma and treated vehicle, ibrutinib, and masitinib for 2 months. The dotted lines were designed to highlight the tumor. (J) MYC expression in blood samples isolated from animals untreated or treated with ibrutinib or masitinib (n = 5 per group). P values were calculated using 2-tailed Student t test. Significant changes between untreated and ibrutinib or masitinib-treated mice are labeled with *P ≤ .05. (K) Survival analysis of animals bearing ABC-lymphoma (patient 13796) untreated or treated with ibrutinib (12 mg/kg per day IP) or masitinib (50 mg/kg per day IP).
Next, we decided to compare the response to ibrutinib or masitinib treatment in an aggressive de novo diagnosed primary patient sample characterized by DH MYC-BCL2 and resistant to available treatments (patient 69487) and a sample of a patient with DLBCL relapsed after treatment with the standard combination of rituximab and chemotherapies and classified as ABC based on expression and IHC analyses (patient 13796). CD20- and CD19-positive B cells were isolated from patient 69487 and were engineered with a luciferase reporter to monitor cell engraftment and tumor progression (Figure 6E). We systemically inoculated 1 million cells into NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) animals; 6 days after injection, when the tumors were expanding (supplemental Figure 5C), we started the treatment with masitinib (n = 13 control and n = 13 masitinib 50 mg/kg IP) or ibrutinib (n = 5 12 mg/kg IP). Strikingly, although DH lymphoma cells were resistant to ibrutinib, the administration of masitinib significantly (P < .0001) limited the accumulation and expansion of tumorigenic B cells (Figure 6F-G) and prolonged the lifespan of the animal harboring DH lymphomas (supplemental Figure 5D) (control, n = 5 [median survival = 37 days]; ibrutinib, n = 5 [median survival = 37 days]; masitinib, n = 5 [median survival = 47 days]). Moreover, we observed that MYC expression in circulating human DLBCL cells correlated with tumor growth, response to masitinib, and resistance to ibrutinib (supplemental Figure 5E). We collected blood from control and treated animals 14, 22, 36, and 44 days after tumor cells engraftment, and we isolated RNA after lysis of the red blood cells. MYC mRNA was undetectable until 22 days after the injection of tumor cells, and it was then progressively accumulating at days 36 and 44 in both the control and ibrutinib-treated animals, anticipating the limited therapeutic benefit of ibrutinib for this patient. Conversely, MYC mRNA level did not significantly change over the course of the treatment with masitinib and, at the last time point, was lower than that in animals treated with ibrutinib or control (supplemental Figure 5E; supplemental Table 8).
For a second patient-derived xenograft model based on ABC-DLBCL cells (patient 13796), we could not engineer the cells with luciferase; thus, we monitored the response to treatment by ultrasound and MYC expression in blood samples (Figure 6H). We inoculated 106 cells through the tail vein, randomized the animals, and started the treatment 1 week after injection. Different from the DH lymphomas, these tumors grew slowly, and MYC was detectable only in the blood of the control group 41 days after injection, but not in ibrutinib- or masitinib-treated animals (supplemental Table 9). Sixty to 70 days after tumor engraftment, we collected blood and assessed the tumor growth by ultrasound. Importantly, we observed a striking difference between treated and untreated animals. Tumor cells were engrafted mainly in the liver, and the control animals presented multiple large masses clearly visible by ultrasound, whereas animals treated with ibrutinib or masitinib had small tumors (Figure 6I; supplemental Figure 5F). We autopsied 1 animal per group and confirmed the differences detected by ultrasound (Figure 6I). Importantly, the response to the treatment was well correlated with a decrease in MYC expression (Figure 6J). Now, we are continuing the treatment to monitor the long-term response or appearance of resistance to treatment, whereas animals treated with the vehicle already reached the end point of the experiment because of the massive tumor burden (supplemental Figure 5F; Figure 6K). During these trials, we observed no loss of weight or alopecia in both NOD-SCID and NSG animals treated with masitinib (supplemental Figure 5G-H), indicating that treatment with masitinib was well tolerated. Hence, masitinib effectively inhibits the proliferation and expansion of aggressive DLBCL lymphoma in preclinical models, and changes in MYC expression levels could be used as a marker to anticipate response to the treatment.
Discussion
BCR signaling is essential for the survival of DLBCL independent of its chronic or tonic activation.23,24 We showed that stimulation of BCR induces activation of multiple downstream targets in all the DLBCL primary samples we tested, independent of their subtype. Nonetheless, differential dependency on BTK favors a response (eg, in ABC-DLBCL) or resistance (eg, in GCB-DLCBL) to BTK selective inhibition.
In this study, we demonstrated that inhibition of BTK in patients who are resistant to ibrutinib changes tumor-signaling dependencies and promotes MYC upregulation, as consequence of a signal imbalance downstream of BCR. Thus, inhibition of BTK in resistant tumors not only is therapeutically ineffective but may also increase tumor aggressiveness. These results underlined the importance of selecting patients who are candidates for ibrutinib treatment and carefully monitoring their response to therapy. Along these lines, we found that MYC expression was detectable in liquid biopsies and that it was correlated with tumor burden and response to ibrutinib treatment, suggesting that it could represent a marker to monitor the therapeutic response.
Pharmacological inhibition of BTK only blocks 1 branch of BCR signaling, and inactivation of 1 arm is compensated by others. Importantly, we found that tumors resistant to BTK blockade become dependent on PI3K/AKT parallel signaling, exposing a new cancer vulnerability that could be therapeutically exploited.
To limit the activation of parallel pathways downstream of the BCR, we proposed to target the root of its signaling. LYN is the most upstream kinase responsible for the initial branching of BCR signaling. Genetic inactivation of LYN did not hamper lymphoma cell proliferation, indicating that LYN-specific targeted therapy would not be effective in DLBCL. LYN activity can be compensated by its paralogs FYN and BLK; indeed, only the triple knockout of these kinases was detrimental to lymphoma cells. Thus, it is necessary to use inhibitors that block the activity of multiple kinases activated by BCR to bypass their redundancy. We found that multitarget inhibitors such as masitinib, bosutinib, and dasatinib can block several SRC kinases concurrently and were therapeutically effective in lymphoma cells in vitro, irrespective of their molecular subtypes (ABC or GCB). Importantly, we demonstrated the effectiveness of masitinib in patient-derived xenografts models of both ABC-DLCBL and DH MYC-BCL2 lymphoma, the latter derived from an aggressive tumor that is often resistant to currently available therapies. Both lymphoma subtypes exhibited high sensitivity to masitinib accompanied by MYC downregulation. Different from the targeted inhibition of BTK, the simultaneous blockade of multiple SRC kinases broadens the possibility of treatment with masitinib to any diagnosed DLBCL or aggressive B-cell lymphoma.
Finally, in this study, we directly measured signaling changes induced by pharmacological agents in primary patient samples, highlighting the possibility of anticipating treatment response. To guide therapeutic decision-making, it is critical not only to screen patients for molecular features but also to monitor the response to therapy during treatment. As shown by our work, preclinical studies in primary samples outline the possibility to monitor the activity of multiple signaling pathways in response to therapies, thus guiding the design of alternative therapeutic approaches.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the École Polytechnique Fédérale de Lausanne research animal, flow cytometry, histology, and sequencing facilities; the protein modeling facility of the University of Lausanne for the support in molecular modeling; and Roy Combe for the help with ultrasound measurements.
This work is supported by the Swiss Institute for Experimental Cancer Research Foundation (E.O.), the Swiss National Science Foundation (E.O. and M.T.) and Swiss Cancer League (E.O. and M.T.), and the Emma Muschamp Foundation (M.T). G.C. is supported by the Giorgi-Cavaglieri Foundation, and L.B. and M.T. were supported by Helmut Horten foundation.
Authorship
Contribution: E.B. was involved in the experimental design and performed the majority of the experiments; N.K. performed in vitro and in vivo experiments; E.D. helped with the characterization of B cells; D.T. helped with analysis of gene expression data; M.C.D. helped with in vivo treatments; L.B. and M.T. helped with the analysis of BCR signaling; A.L.C. and M.A.M. provided histological and molecular characterization of the primary samples; O.M. and V.Z. performed protein-ligand docking analysis; G.C. supervised the analysis of expression data and drug screen; and E.O. designed the study, supervised the experiments and wrote the manuscript with comments from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elisa Oricchio, Swiss Institute for Experimental Cancer Research, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; e-mail: elisa.oricchio@epfl.ch.

![Figure 2. Treatment with ibrutinib changes MYC expression in cells sensitive and resistant to ibrutinib. (A-B) Percentage of viable cells in the indicated lymphoma cell lines treated with DMSO or 0.1 or 0.5 μM ibrutinib for 72 hours. Each cell line was analyzed in triplicate, and data are shown as a bar graph corresponding to the mean ± SD. (C) Western blot analysis of pBTK levels in GCB and ABC cells after DMSO or ibrutinib treatment (0.5 μM, 6 hours) and BCR stimulation (20 μg/mL F[ab']2 anti-human immunoglobulin M, 3 min). (D-G) Phosphoflow cytometry analysis and quantification of CD19 (Tyr 531) (D-E) and GSK3β (Ser 9) (F-G) in stimulated lymphoma cells pretreated with DMSO or ibrutinib (0.5 μM) for 6 hours, normalized to the stained, unstimulated controls. (H) MYC expression in the indicated ibrutinib-resistant cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates. Data have been normalized to the DMSO-treated control and are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib-treated cells were labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. (I) Western blot analysis of the indicated ibrutinib-resistant cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. (J) MYC expression in the indicated ibrutinib-sensitive cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Each cell line was analyzed in 3 biological replicates. Data have been normalized to the DMSO-treated control and are shown as a bar graph corresponding to the mean ± SD. P values were calculated using 2-tailed Student t test. Significant changes between DMSO-treated and ibrutinib-treated cells were labeled with *P ≤ .05; **P ≤ .01; ***P ≤ .001. (K) Western blot analysis of the indicated ibrutinib-sensitive cell lines treated with DMSO or ibrutinib (0.5 μM) for 24 or 48 hours. Signal quantification was performed using Image Studio Lite and normalized to the DMSO-treated control. (L) Western blot analysis of MYC and BTK in isogenic GCB cell lines WT or BTK KO. (M) Heat map indicating the expression levels (z-scores) of 10 MYC target genes in WSU-DLCL2 cells WT or BTK KO.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/21/10.1182_blood-2017-10-809210/4/m_blood809210f2.jpeg?Expires=1767752351&Signature=uxQimsf0AdZDUDYyvlHgNoaiu0xd9EdSoeRFyF~uKLCuWLegNAUNb5fHsavfgG2hJWguImKOScBBjLYGVPy1UNi8P11boSgIqZ55yvhmlbnjmo32-ThDth7kLIJruLboQPGpWHcFoJA0s5eX4OjoG4U2rpNdtW5-9kVV~d6FNuIcg4oO53p2Wz~OhQKH0ge9nRXy-CCAMS5Gkhiv7zqOidciWehwwOTVy8smSMdCA6L8SmIRamM0sfsuI3psR7F3jvLz3rAGGIEYB3hU1r9IhPMdK~m7WNtanUOwyPivIMTKCpzsTjaruke23cujqjL7-VHx2UuOSssOpQFbUMte3w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal