Key Points
CD3γ-deficient patients manifest T-cell phenotypic and functional defects that are especially prominent in Treg cells.
The peripheral T-cell repertoire of CD3γ-deficient patients is restricted, with molecular signatures of self-reactivity.
Abstract
Integrity of the T-cell receptor/CD3 complex is crucial for positive and negative selection of T cells in the thymus and for effector and regulatory functions of peripheral T lymphocytes. In humans, CD3D, CD3E, and CD3Z gene defects are a cause of severe immune deficiency and present early in life with increased susceptibility to infections. By contrast, CD3G mutations lead to milder phenotypes, mainly characterized by autoimmunity. However, the role of CD3γ in establishing and maintaining immune tolerance has not been elucidated. In this manuscript, we aimed to investigate abnormalities of T-cell repertoire and function in patients with genetic defects in CD3G associated with autoimmunity. High throughput sequencing was used to study composition and diversity of the T-cell receptor β (TRB) repertoire in regulatory T cells (Tregs), conventional CD4+ (Tconv), and CD8+ T cells from 6 patients with CD3G mutations and healthy controls. Treg function was assessed by studying its ability to suppress proliferation of Tconv cells. Treg cells of patients with CD3G defects had reduced diversity, increased clonality, and reduced suppressive function. The TRB repertoire of Tconv cells from patients with CD3G deficiency was enriched for hydrophobic amino acids at positions 6 and 7 of the CDR3, a biomarker of self-reactivity. These data demonstrate that the T-cell repertoire of patients with CD3G mutations is characterized by a molecular signature that may contribute to the increased rate of autoimmunity associated with this condition.
Introduction
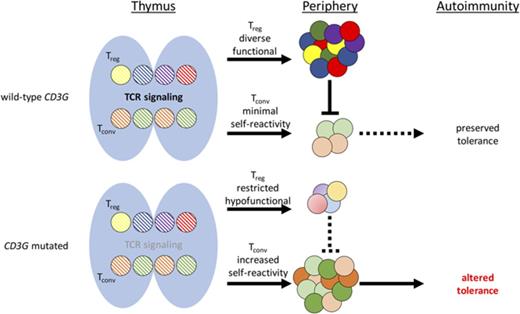
Integrity of the T-cell receptor/CD3 (TCR/CD3) complex is crucial for T-cell maturation. In particular, the strength of TCR signaling plays a critical role in governing positive and negative selection in the thymus as well as responses of effector and regulatory T (Treg) cells in the periphery. Before reaching the membrane, TCRα/β heterodimers associate with 3 invariant dimers (CD3δ/ε, CD3γ/ε, and CD3ζ/ζ) that compose the CD3 complex.1 Following localization on the cell surface, the CD3 proteins convert ligand recognition by α/β TCR chains into intracellular signals.2
Both in humans and in mice, genetic defects that cause complete absence of CD3δ or CD3ε chain expression lead to a block in the development of TCRαβ+ T cells and are a cause of severe combined immune deficiency.3-5 Human CD3ζ deficiency is characterized by a reduced number of circulating T cells that are nonfunctional and display a restricted T-cell repertoire, thereby causing severe immunodeficiency.6 Although a severe block in T-cell development is also observed in CD3γ-deficient mice,7 it has been shown that the loss of CD3γ protein in humans allows the development of polyclonal T cells with impaired, but not abolished, TCR/CD3 signaling, and is associated with a milder clinical phenotype characterized by a variable degree of susceptibility to infections and the frequent occurrence of autoimmune manifestations.8-10
A similar phenotype has been also reported in patients and mice with hypomorphic mutations in genes that encode for signaling molecules downstream of the TCR/CD3 complex.11,12 Altogether, these observations are consistent with the notion that TCR signaling strength plays a critical role both in T-cell development and function and in establishing and maintaining central and peripheral tolerance.
To gain novel insights into how CD3G mutations in humans affect T-cell development and homeostasis, we have analyzed TCR diversity and composition, T-cell proliferation, and Treg number and function in 6 patients with CD3γ deficiency and in healthy controls.
Methods
Human subjects
Deidentified patients’ clinical and immunologic data were provided by an international network of physicians in the United States, Europe, and Asia. All human subject samples were consented under protocols approved by the institutional review boards at the participating institutions. The study was approved by the institutional review board at Boston Children's Hospital (protocol 0409113R) and at the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda (protocol 16-I-N139). The study met the institutional review board standards for ethical conduct of research with human subjects.
Flow cytometry/fluorescent activated cell sorting
Peripheral blood mononuclear cells (PBMCs) were processed using Ficoll (GE Healthcare, Malborough, MA) to form a single cell suspension and then stained with the following monoclonal antibodies directed against cell surface antigens: CD4-AlexaFluor700, CD8a-PE/Dazzle594, CD19-PerCPCy5.5, CD127-PECy7, and CD25-PE (all from Biolegend, San Diego, CA). Intranuclear Foxp3-eFluor450 (eBioscience, San Diego, CA) or Ki67 (Becton Dickinson San Jose, CA) staining was performed using the Foxp3 staining buffers (eBioscience). For further characterization of Treg cells, PBMC were also stained with monoclonal antibodies against CTLA-4 (clone BNI3, eBioscience), ICOS/CD278 (clone DX29, eBioscience), and HELIOS (clone 22F6, eBioscience), along with CD4 and FOXP3. For intracellular staining, fixation/permeabilization buffer (eBioscience) was used according to the manufacturer’s instructions. Upon washing, cells were analyzed by flow cytometry using LSRFortessa, and results were analyzed using FlowJo software, version 8.8.6 (Tree Star Ashland, OR). In parallel, PBMC were stained with CD3-eFluor450 (OKT3, eBioscience), CD4-FITC, CD8a-APC, TCRαβ-APC (clone IP26), mouse IgG1-eFluor450, and mouse IgG1-APC (all from Biolegend), followed by cell sorting using FACSAria (Becton Dickinson).
Cell proliferation assay
PBMC were isolated to form a single cell suspension and then stained with carboxyfluorescein succinimidyl ester (CFSE) (Thermo Fisher, Carlsbad, CA) at a concentration of 5 μM/mL for 10 minutes at 37°C in phosphate-buffered saline. Cells where then washed, resuspended in RPMI containing 10% fetal bovine serum, plated in a 48-well plate at a concentration of 5 × 105 cells per well, and then cultured with either medium alone (no stimulation), 5 μg/mL phytohemagglutinin (PHA; Sigma Aldrich, St. Louis, MO), or 100 ng/mL anti-CD3 (eBioscience) either alone or in combination with 100 ng/mL anti-CD28 (eBioscience) for 4 days. For patient 6, these standard doses were titrated by log increments to create the dose-response curves. The cells were then stained for flow cytometry as indicated previously, and proliferation was assessed by CFSE dilution as described.13
Treg suppression assay
For evaluating Treg suppressive potency, CD4+ cells were enriched by magnetic bead separation (Miltenyi Biotec, Germany) followed by fluorescent activated cell sorting into the following subsets: CD4+ CD25hi CD127low cells (Treg cells) and CD4+ CD25low CD127hi T effector (Teff) cells. In each experiment, CD4+ Teff cells were stained using Cell Trace Violet (Thermo Fisher) according to the manufacturer’s instructions. Treg and Teff cells were then mixed at the indicated ratios based on a conventional CD4+ (Tconv) count of 5000 cells per well, then stimulated with anti-CD2/CD3/CD28 beads (Treg Suppression Inspector, Miltenyi Biotec) at a 1:1 Tconv:bead ratio in complete RPMI media for 4 days. In this assay, T-cell proliferation is indicated by dilution of Cell Trace Violet. The relative Treg suppressive activity was determined by plotting the percentage of dividing cells at each Treg:Teff ratio.
High throughput sequencing (HTS) of TCRβ
PBMCs were collected and sorted into CD3+ CD4+ CD8− CD25hi CD127low Treg, CD3+ CD4+ CD8−CD25low CD127hi Tconv, and CD3+ CD8+ CD4− T-cell populations using FACSAria cell sorter (Becton Dickinson). The TCRβ (TRB) rearranged genomic products were amplified by multiplex polymerase chain reaction (PCR), using DNA as the template (Adaptive Biotechnologies Seattle, WA). To amplify all possible VDJ combinations, 52 forward primers for the TRBV gene segments and 13 reverse primers to the TRBJ gene segments were used. Adaptive Biotechnologies uses assay-based and computational techniques to minimize PCR amplification bias. The assay is quantitative, and the frequency of a given TRB sequence is representative of the frequency of that clonotype in the original sample. The PCR products were sequenced using the Illumina HiSeq platform. Custom algorithms were used to filter the raw sequences for errors and align the sequences to reference genome sequences. Subsequently, the data were analyzed using the ImmunoSeq set of online tools. The frequency of productive and nonproductive TRB rearrangements was analyzed within both unique and total TRB sequences obtained from sorted T-cell subsets. Distribution of the frequency of individual clonotypes (including TRBV to TRBJ pairing) and use of individual TRBV genes were analyzed within unique sequences. Heat map representation of the frequencies of individual TRBV to TRBJ gene pairs and sequence overlap was produced using Morpheus software (https://software.broadinstitute.org/morpheus/). TCR self-reactivity was analyzed by analyzing amino acid composition and hydrophobicity at positions 6 and 7 of the TRB CDR3, as previously described.14 TRB sequences from patients and controls are available at: https://doi.org/10.21417/B7XS7J.
Statistical analysis
The number and percentage of cells and of TRB HTS reads were first analyzed and found to follow a Gaussian distribution. The differences between groups were analyzed using either Student t test (for individual comparisons with gaussian distributions), Mann-Whitney test (for individual comparisons with nongaussian distributions), or 2-way analysis of variance (for multiple comparisons) with statistical significance indicating a 95% confidence interval >0, *P < .05, **P < .01, ***P < .001, and ****P < .0001. All statistical analysis was performed using Prism Software.
Results
Patients’ molecular, clinical, and immunological features
Six patients carrying biallelic CD3G mutations were studied; their molecular, clinical, and laboratory features are reported in Table 1. Five patients were homozygous, and 1 patient was compound heterozygous for CD3G mutations. Patient 2 (P2) and P3, and P4 and P5 were 2 pairs of siblings. All patients had evidence of autoimmunity, which was more severe in P1 and P4 (supplemental Table 1, available on the Blood Web site). A history of infections was present in 4 patients, and was more severe in P1, P4, and P6. Significant variability of the immunological profile was observed. In particular, a low CD4+ cell count was observed in P4, and decreased CD8+ cell counts were detected in P2, P4, and P6. All patients had a reduced proportion of naïve T cells and increased percentage of memory CD4+ and CD8+ cells, including accumulation of a large proportion of CD45RA+ CCR7− TEMRA “exhausted” CD8+ T cells in 5 (P1-P5) (Table 1). Low IgG serum levels were present in P1, P4, and P6.
Clinical and immunological phenotype of CD3G-mutated patients
| Patient . | P1 . | P2 . | P3 . | P4 . | P5 . | P6 . |
|---|---|---|---|---|---|---|
| Age at testing | 27 mo | 19 y | 6 y | 15 y | 23 y | 34 y |
| CD3G mutation (allele 1) | c.1A>G | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.1A>G |
| CD3G mutation (allele 2) | c.1A>G | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.80-1G>C |
| CD3, cells/μL | 2018 (850-4300) | 630 (850-3200) | 2800 (2400-8300) | 380 (850-3200) | 1200 (570-2400) | 1051 (850-3200) |
| CD4, cells/μL | 627 (500-2700) | 520 (400-2100) | 1700 (1300-7100) | 210 (400-2100) | 800 (430-1800) | 842 (400-2100) |
| Of which are naïve, % | 39.4 | 19.3 | 41.3 | 5.4 | 13.0 | 3.1 |
| CM, % | 40.3 | 25.5 | 20.7 | 61.7 | 67.2 | 87 |
| EM, % | 19.8 | 53.5 | 34.4 | 31.5 | 19.7 | 9.5 |
| EMRA, % | 0.5 | 1.7 | 3.6 | 1.5 | 0.1 | 0.1 |
| CD8, cells/μL | 1139 (200-1800) | 40 (300-1300) | 822 (400-4100) | 116 (300-1300) | 270 (210-1200) | 200 (300-1300) |
| Of which are naïve, % | 7.4 | 7.7 | 26.0 | 5.3 | 11.3 | 5.0 |
| CM, % | 1.6 | 0.7 | 1.2 | 3.0 | 9.0 | 45.5 |
| EM, % | 41.4 | 57.5 | 44.9 | 55.4 | 53.5 | 44.7 |
| EMRA, % | 49.6 | 34.1 | 27.9 | 36.4 | 26.1 | 4.7 |
| CD19, cells/μL | 1988 (180-1300) | 250 (120-740) | 170 (110-7700) | 135 (120-740) | 800 (91-610) | 195 (120-740) |
| CD16/56, cells/μL | 945 (61-510) | 920 (92-1200) | 1500 (71-3500) | 370 (92-1200) | 500 (92-1200) | 137 (74-620) |
| IgG, mg/dL | 338 (407-1009) | 1240 (768-1632) | 881 (331-1164) | 491 (768-1632) | 1660 (768-1632) | 632 (768-1632) |
| IgA, mg/dL | 20 (14-122) | 180 (68-408) | 18 (14-105) | 6.6 (68-408) | 165 (68-408) | 364 (68-408) |
| IgM, mg/dL | 41 (46-160) | 110 (35-263) | 213 (41-164) | 50.4 (35-263) | 219 (35-263) | 59 (35-263) |
| IgE, kU/L | <1 (≤199) | 239 (≤696) | 6 (≤97) | 6 (≤696) | 6 (≤214) | ND |
| Infections | Klebsiella pneumoniae, MSSA, severe EBV | Acute bronchitis (×3) | none | RRTI, bronchiectasis | none | Soft tissue abscesses, viral meningitis |
| Autoimmunity | Autoimmune enteropathy, GLILD, AIHA | Hypothyroidism | Hypothyroidism | Hypothyroidism, AIHA, ITP, nephropathy, ALPS | Thyroiditis | Vitiligo, hypothyroidism |
| Skin rash | No | Atopic dermatitis | No | No | Atopic dermatitis | Atopic dermatitis |
| Patient . | P1 . | P2 . | P3 . | P4 . | P5 . | P6 . |
|---|---|---|---|---|---|---|
| Age at testing | 27 mo | 19 y | 6 y | 15 y | 23 y | 34 y |
| CD3G mutation (allele 1) | c.1A>G | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.1A>G |
| CD3G mutation (allele 2) | c.1A>G | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.80-1G>C | c.80-1G>C |
| CD3, cells/μL | 2018 (850-4300) | 630 (850-3200) | 2800 (2400-8300) | 380 (850-3200) | 1200 (570-2400) | 1051 (850-3200) |
| CD4, cells/μL | 627 (500-2700) | 520 (400-2100) | 1700 (1300-7100) | 210 (400-2100) | 800 (430-1800) | 842 (400-2100) |
| Of which are naïve, % | 39.4 | 19.3 | 41.3 | 5.4 | 13.0 | 3.1 |
| CM, % | 40.3 | 25.5 | 20.7 | 61.7 | 67.2 | 87 |
| EM, % | 19.8 | 53.5 | 34.4 | 31.5 | 19.7 | 9.5 |
| EMRA, % | 0.5 | 1.7 | 3.6 | 1.5 | 0.1 | 0.1 |
| CD8, cells/μL | 1139 (200-1800) | 40 (300-1300) | 822 (400-4100) | 116 (300-1300) | 270 (210-1200) | 200 (300-1300) |
| Of which are naïve, % | 7.4 | 7.7 | 26.0 | 5.3 | 11.3 | 5.0 |
| CM, % | 1.6 | 0.7 | 1.2 | 3.0 | 9.0 | 45.5 |
| EM, % | 41.4 | 57.5 | 44.9 | 55.4 | 53.5 | 44.7 |
| EMRA, % | 49.6 | 34.1 | 27.9 | 36.4 | 26.1 | 4.7 |
| CD19, cells/μL | 1988 (180-1300) | 250 (120-740) | 170 (110-7700) | 135 (120-740) | 800 (91-610) | 195 (120-740) |
| CD16/56, cells/μL | 945 (61-510) | 920 (92-1200) | 1500 (71-3500) | 370 (92-1200) | 500 (92-1200) | 137 (74-620) |
| IgG, mg/dL | 338 (407-1009) | 1240 (768-1632) | 881 (331-1164) | 491 (768-1632) | 1660 (768-1632) | 632 (768-1632) |
| IgA, mg/dL | 20 (14-122) | 180 (68-408) | 18 (14-105) | 6.6 (68-408) | 165 (68-408) | 364 (68-408) |
| IgM, mg/dL | 41 (46-160) | 110 (35-263) | 213 (41-164) | 50.4 (35-263) | 219 (35-263) | 59 (35-263) |
| IgE, kU/L | <1 (≤199) | 239 (≤696) | 6 (≤97) | 6 (≤696) | 6 (≤214) | ND |
| Infections | Klebsiella pneumoniae, MSSA, severe EBV | Acute bronchitis (×3) | none | RRTI, bronchiectasis | none | Soft tissue abscesses, viral meningitis |
| Autoimmunity | Autoimmune enteropathy, GLILD, AIHA | Hypothyroidism | Hypothyroidism | Hypothyroidism, AIHA, ITP, nephropathy, ALPS | Thyroiditis | Vitiligo, hypothyroidism |
| Skin rash | No | Atopic dermatitis | No | No | Atopic dermatitis | Atopic dermatitis |
Reference values for age are shown in parentheses.
AIHA, autoimmune hemolytic anemia; ALPS, autoimmune lymphoproliferative syndrome; CM, central memory (CD45RA− CCR7+); EBV, Epstein-Barr virus; EM, effector memory (CD45RA− CCR7−); EMRA, effector memory CD45RA+ (CD45RA+ CCR7−; GLILD, granulomatous lymphocytic interstitial lung disease; ITP, idiopathic thrombocytopenic purpura; MSSA, methicillin-sensitive Staphylococcus aureus; ND, no data; RRTI, recurrent respiratory tract infections.
CD3γ is essential for optimal expression of surface CD3/TCRαβ complex
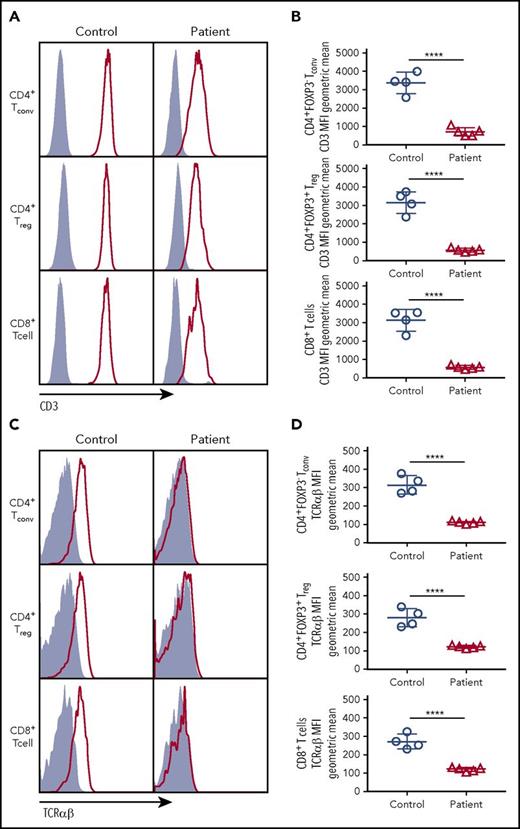
To determine the impact of CD3G mutations on the expression of the TCR/CD3 complex, we analyzed CD8+ T cells, CD4+ Tconv, and CD4+ Treg cells from patients P1 to P5 and controls, and found that expression of both CD3 (as indicated by staining with a monoclonal antibody specific for the CD3ε chain) and TCRαβ was markedly reduced in each of these subsets in all patients with CD3G mutations (Figure 1). These findings confirm and extend previous observations10,15-17 and demonstrate that CD3γ is required to maintain optimal levels of surface TCRαβ and CD3 complex expression in all T-cell subsets.
CD3 and TCRαβ expression on the surface of T-cell subsets. (A) Expression of CD3 (red line) or isotype (solid gray) on the surface of CD4+ FOXP3− Tconv, CD4+FOXP3+ Treg, and CD8+ T cells. (B) Cumulative data of geometric mean for mean fluorescence intensity of CD3 surface expression among Tconv, Treg, or CD8+ cells. (C) Expression of TCRαβ (red line) or isotype (solid gray) on the surface of Tconv, Treg. and CD8+cells. (D) Cumulative data of geometric mean for mean fluorescence intensity of TCRαβ surface expression among Tconv, Treg, and CD8+ cells. (B,D) Five patients (P1-P5) were studied.
CD3 and TCRαβ expression on the surface of T-cell subsets. (A) Expression of CD3 (red line) or isotype (solid gray) on the surface of CD4+ FOXP3− Tconv, CD4+FOXP3+ Treg, and CD8+ T cells. (B) Cumulative data of geometric mean for mean fluorescence intensity of CD3 surface expression among Tconv, Treg, or CD8+ cells. (C) Expression of TCRαβ (red line) or isotype (solid gray) on the surface of Tconv, Treg. and CD8+cells. (D) Cumulative data of geometric mean for mean fluorescence intensity of TCRαβ surface expression among Tconv, Treg, and CD8+ cells. (B,D) Five patients (P1-P5) were studied.
CD3γ deficiency affects T-cell proliferation in response to mitogen stimulation
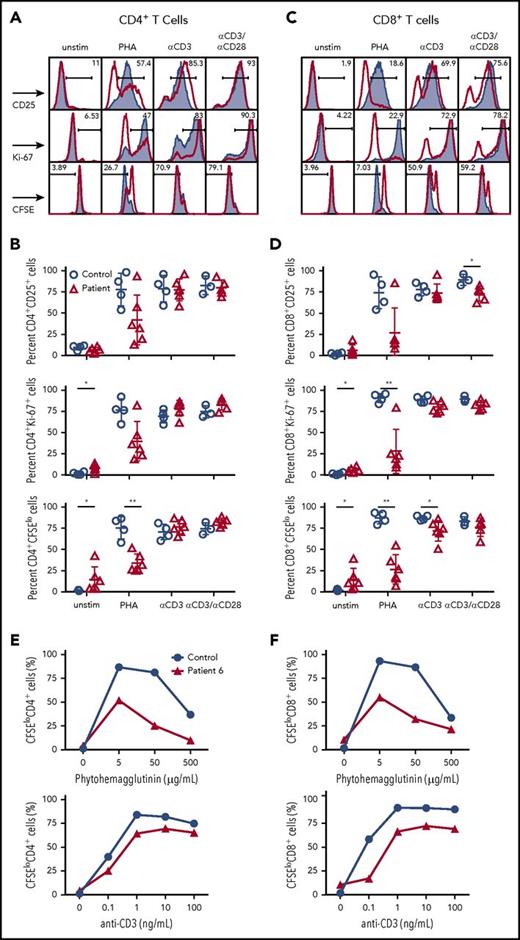
Next, we analyzed whether the impaired surface expression of CD3/TCRαβ complex in T cells from CD3G-mutated patients would also affect cellular activation and proliferation. When stimulated with phytohemagglutinin (PHA), both CD4+ and CD8+ T cells in CD3γ-deficient patients demonstrated reduced cellular division (as shown by CFSE dilution); a similar trend was observed when analyzing cellular activation (by staining for CD25) and proliferation (by Ki-67 staining) (Figure 2A-D).
T-cell activation and cell division following mitogen stimulation. (A) Representative plots and (B) cumulative percentage of (top) CD25 expression, (middle) Ki67 expression, and (bottom) CFSE staining in CD4+ T cells from controls (solid gray) or CD3G patients (red) in unstimulated samples or following activation with PHA, anti-CD3, or anti-CD3/CD28. (C) Representative plots and (D) cumulative data for (top) CD25 expression, (middle) Ki67 expression, and (bottom) CFSE staining in CD8+ T cells from controls (solid gray) or patients (red) among the indicated groups. (A-D) Response to anti-CD3+anti-CD28 was studied in 5 patients (P1-P5) because not enough cells were available from P6. Dose titration curve of CFSE dilution among (E) CD4+ and (F) CD8+ T cells following stimulation with PHA (top) or anti-CD3 (bottom) for patient P6 (red line) and control sample (blue line). Unstim, unstimulated.
T-cell activation and cell division following mitogen stimulation. (A) Representative plots and (B) cumulative percentage of (top) CD25 expression, (middle) Ki67 expression, and (bottom) CFSE staining in CD4+ T cells from controls (solid gray) or CD3G patients (red) in unstimulated samples or following activation with PHA, anti-CD3, or anti-CD3/CD28. (C) Representative plots and (D) cumulative data for (top) CD25 expression, (middle) Ki67 expression, and (bottom) CFSE staining in CD8+ T cells from controls (solid gray) or patients (red) among the indicated groups. (A-D) Response to anti-CD3+anti-CD28 was studied in 5 patients (P1-P5) because not enough cells were available from P6. Dose titration curve of CFSE dilution among (E) CD4+ and (F) CD8+ T cells following stimulation with PHA (top) or anti-CD3 (bottom) for patient P6 (red line) and control sample (blue line). Unstim, unstimulated.
Similar, albeit less pronounced, defects in division were observed in CD8+, but not in CD4+ T cells, from CD3G-mutated patients upon in vitro stimulation with anti-CD3. Stimulation with both anti-CD3 and anti-CD28 resulted in overall equivalent activation, proliferation, and cell division in both CD4+ and CD8+ T cells from patients and controls (Figure 2A-D).
Next, we interrogated whether the impaired cell activation and proliferation observed in response to optimal concentrations of PHA (5 μg/mL) could be overcome by using higher PHA concentrations. Peripheral blood CD8+ and CD4+ T cells from P6 and a healthy control were stimulated in vitro with increasing concentrations of PHA. As shown in Figure 2E-F, a lower percentage of dividing CD8+ and CD4+ T cells was detected in P6 than in the healthy control at any PHA concentration. A subtle defect was also observed on stimulation with various concentrations of anti-CD3. Finally, the peak of cell division was reached at the same concentration of PHA or anti-CD3 both in the healthy control and in P6. Taken together, these results demonstrate the impaired surface expression of the TCR/CD3 complex in CD3γ-deficient T cells is associated with reduced cellular proliferation after mitogen stimulation, and that this defect cannot be circumvented by increasing the strength of the stimulating condition.
Treg cells from patients with CD3γ deficiency have decreased suppressive activity
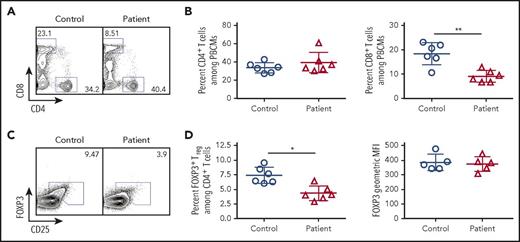
Given the importance of TCR signaling in Treg development and homeostasis,18-20 we hypothesized that this subpopulation of cells would be disproportionately affected by reduced TCR/CD3 signaling in patients with CD3G mutations. Among CD4+ T cells, the proportion of CD25hi FOXP3+ Treg cells was significantly lower in CD3G-mutated patients than in controls. However, levels of FOXP3 protein expression were similar in Treg cells from patients and controls (Figure 3). Other phenotypic markers of Treg cells (CTLA-4, HELIOS, ICOS) were equally expressed both in patient- and in control-derived Treg cells (supplemental Figure 2).
Flow cytometric analysis of peripheral blood lymphocyte populations in patients with CD3G mutations. (A) Representative flow cytometry plots for circulating CD4+ and CD8+ T lymphocytes in controls and CD3G-mutated patients. (B) Percentage of CD4+ (left) and CD8+ (right) T cells among PBMCs for control and CD3G-mutated patients. (C). Representative flow cytometry for Treg identification by FOXP3 and CD25 expression. (D) Percentage (left) of FOXP3+ Treg cells and FOXP3 mean fluorescence intensity (right) for controls and patients.
Flow cytometric analysis of peripheral blood lymphocyte populations in patients with CD3G mutations. (A) Representative flow cytometry plots for circulating CD4+ and CD8+ T lymphocytes in controls and CD3G-mutated patients. (B) Percentage of CD4+ (left) and CD8+ (right) T cells among PBMCs for control and CD3G-mutated patients. (C). Representative flow cytometry for Treg identification by FOXP3 and CD25 expression. (D) Percentage (left) of FOXP3+ Treg cells and FOXP3 mean fluorescence intensity (right) for controls and patients.
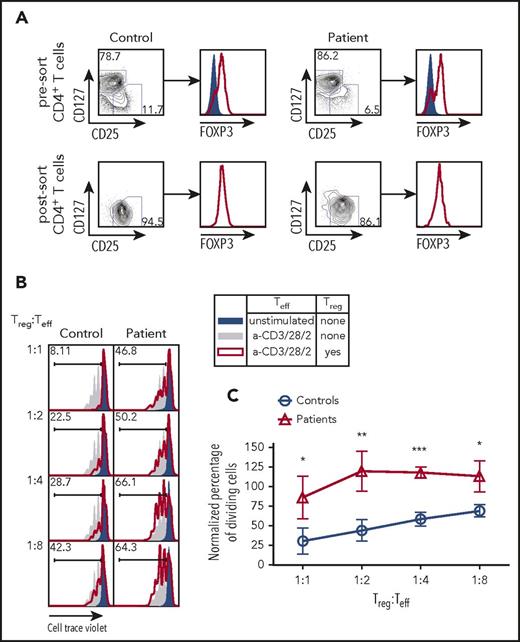
Next, we sought to determine whether the impairment of TCR/CD3 signaling observed in CD3γ-deficient T cells would also affect the capacity of Treg cells to suppress the activation of Tconv cells. CD4+ T cells from 4 patients (P2-P5) and controls were sort-purified into CD25hi CD127low Treg and CD25low CD127hi Teff cells (Figure 4A). The Teff cells from controls were assayed for cell division following coculture with varying ratios of Treg cells isolated from either controls or patients. Within each experiment, the percentage of dividing cells was normalized to the percentage of dividing Teff cells upon stimulation with anti-CD2/CD3/CD28 beads in the absence of Treg cells (Figure 4B; solid gray area). Teff cells cocultured with control Treg cells demonstrated a linear reduction in cell proliferation as the number of Treg cells was uptitrated to a final Treg:Teff cell ratio of 1:1 (Figure 4B-C). By contrast, Treg cells from CD3G-mutated patients completely failed to suppress proliferation of control Teff cells up to a 1:2 Treg:Teff cell ratio; reduced suppressive activity was also detected at a 1:1 Treg:Tconv ratio (Figure 4B-C). Together, these findings demonstrate that Treg cells isolated from patients with CD3G mutations are unable to suppress the proliferation of Teff cells upon stimulation, suggesting that loss of Treg suppressive function may be an important component of altered peripheral T-cell tolerance in patients with CD3γ deficiency.
Treg suppression assay from fluorescent activated cell sorting purified Treg cells in CD3G-mutated patients. (A) CD25hiCD127lo Treg sort purification strategy from control (left) and patient (right) samples. Intranuclear FOXP3 staining was performed on CD25loCD127hi Teff cells (solid blue) and CD25hiCD127lo Treg cells (red line). (B) Representative plots of Cell Trace Violet dilution in experiments in which CD4+ CD25low CD127hi Teff cells from controls were either left unstimulated (solid blue) or stimulated with anti-CD3 + anti-CD28 + anti-CD2, in the absence (solid gray) or presence (red line) of Treg cells from controls (left) or from CD3G-mutated patients (right). (C) Cumulative percentage of dividing cells in Treg suppression assay, normalized to the no Treg group within each individual experiment. Treg function was tested in 4 patients (P2-P5) and 4 controls. Results are expressed as mean ± standard deviation. *P < .05; **P < .01; ***P < .001.
Treg suppression assay from fluorescent activated cell sorting purified Treg cells in CD3G-mutated patients. (A) CD25hiCD127lo Treg sort purification strategy from control (left) and patient (right) samples. Intranuclear FOXP3 staining was performed on CD25loCD127hi Teff cells (solid blue) and CD25hiCD127lo Treg cells (red line). (B) Representative plots of Cell Trace Violet dilution in experiments in which CD4+ CD25low CD127hi Teff cells from controls were either left unstimulated (solid blue) or stimulated with anti-CD3 + anti-CD28 + anti-CD2, in the absence (solid gray) or presence (red line) of Treg cells from controls (left) or from CD3G-mutated patients (right). (C) Cumulative percentage of dividing cells in Treg suppression assay, normalized to the no Treg group within each individual experiment. Treg function was tested in 4 patients (P2-P5) and 4 controls. Results are expressed as mean ± standard deviation. *P < .05; **P < .01; ***P < .001.
Skewed use of TRBV and TRBJ genes and abnormalities of TRB CD3 composition in patients with CD3G mutations
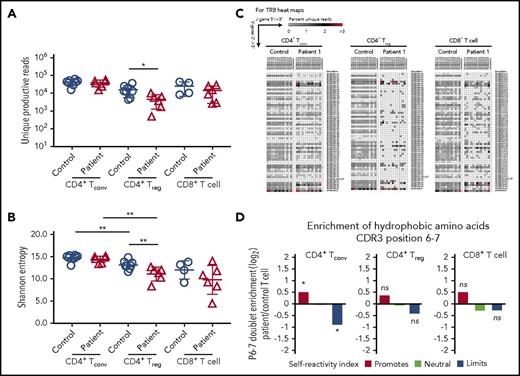
Reduced TCR/CD3 expression and signaling may also affect mechanisms of positive and negative selection in the thymus,21,22 with CD3γ playing an important role in this process.23 To investigate in greater detail how human CD3γ deficiency may alter peripheral T-cell repertoire diversity and composition, we performed HTS to analyze TRB diversity, CDR3 characteristics, and TRB V-J pair gene use in patient T cells. Peripheral blood T-cell subsets were sort-purified into CD8+ T cells, CD4+ CD25hi CD127lo Treg cells, and CD4+ CD25low CD127hi Tconv cells. Upon HTS of TRB rearrangements, there was a ∼log10 reduction in the number of unique productive reads identified in CD4+ Treg cells from CD3γ-deficient patients compared with controls, whereas no differences were observed within CD4+ Tconv and CD8+ T cells (Figure 5A). Consistent with previous studies,24,25 Shannon entropy index revealed equivalent diversity of Treg and Tconv cells in controls samples (Figure 5B). However, Treg cells from CD3γ-deficient patients had reduced diversity compared with autologous Tconv cells and with Treg cells from healthy controls (Figure 5B). There was a trend toward reduced Shannon entropy index for patient-derived CD8+ T cells compared with control CD8+ cells, although the difference was not statistically significant.
HTS analysis of TRB repertoire in CD4+Treg, CD4+Tconv, and CD8+T-cell subsets. (A) Number of unique reads identified in each patient for Treg, Tconv, and CD8+ T cells and (B) Shannon entropy index calculated among unique reads for Treg, Tconv, and CD8+ T cells. (A-B) *P < .05; **P < .01. (C) Representative heat maps depicting Vβ and Jβ gene pairing in unique sequences from (left) CD4+ Tconv, (middle) CD4+ Treg, and (right) CD8+ T cells from controls and patient 1. (D) Hydrophobicity analysis of amino acid residues at positions 6 and 7 of the TRB-CDR3 in (left) CD4+ Tconv, (middle) CD4+ Treg, and (right) CD8+ T cells from CD3G-mutated patients (n = 6) compared with controls (n = 8). *P < 10−4.
HTS analysis of TRB repertoire in CD4+Treg, CD4+Tconv, and CD8+T-cell subsets. (A) Number of unique reads identified in each patient for Treg, Tconv, and CD8+ T cells and (B) Shannon entropy index calculated among unique reads for Treg, Tconv, and CD8+ T cells. (A-B) *P < .05; **P < .01. (C) Representative heat maps depicting Vβ and Jβ gene pairing in unique sequences from (left) CD4+ Tconv, (middle) CD4+ Treg, and (right) CD8+ T cells from controls and patient 1. (D) Hydrophobicity analysis of amino acid residues at positions 6 and 7 of the TRB-CDR3 in (left) CD4+ Tconv, (middle) CD4+ Treg, and (right) CD8+ T cells from CD3G-mutated patients (n = 6) compared with controls (n = 8). *P < 10−4.
Consistent with the reduced diversity, Treg cells from CD3γ-deficient patients demonstrated a marked restriction in the number of unique TRB VJ pairs identified (Figure 5C; supplemental Figure 1). A similar, albeit less pronounced, restriction was present also in CD4+ Tconv and in CD8+ T cells from patients compared with controls (Figure 5C; supplemental Figure 1). This restriction in TRB diversity was driven by skewed usage of both the TRB V and J genes (supplemental Table 1). Interestingly, similar differences were observed in Treg and Tconv cells for individual V and J genes that were under- or overrepresented in patients vs controls (supplemental Table 1). Together, these results indicate that mutations in CD3G result in skewing of the TRB repertoire through alterations in the frequency of V and J gene use, which results in restriction of the overall TCR repertoire.
Because patients with CD3G mutations develop multiple autoimmune manifestations, we sought to further characterize the TRB repertoire for properties of the CDR3 region that may associate with self-reactivity. In particular, the presence of hydrophobic amino acids at positions 6 and 7 of the TRB-CDR3 has been previously shown to promote self-reactivity in both mouse and human TCRs.14 Among Treg cells, there were no significant changes in the relative hydrophobicity of amino acid positions 6 and 7 when comparing CD3G-mutated patients with controls (Figure 5D). However, the CD4+ Tconv cell population from CD3G-mutated patients contained significantly more reads enriched in hydrophobic amino acid residues at positions 6 and 7 of the TRB CDR3, compared with controls (Figure 5D), suggesting enrichment in self-reactive specificities among Tconv cells.
Discussion
Autoimmunity is a frequent complication of primary immune deficiencies. In particular, patients with genetic defects reducing the strength of TCR/CD3 signaling are exquisitely susceptible to the development of autoimmunity.11 The number, diversity, and function of Treg cells have each been demonstrated to be critical factors for sustaining immune tolerance,26-30 and changes in the relative suppressive potency of Treg cells have been demonstrated to lead to autoimmunity in patients and mouse models with autoimmunity.31-36 However, the relative contribution of abnormalities of Treg cell number, diversity, and function to the development of clinical autoimmune disease in patients with genetic defects of TCR signaling remains largely unknown. Herein, we performed a comprehensive analysis of peripheral T-cell number, phenotype, function, and repertoire in patients with disease-causing CD3G mutations. We have demonstrated that genetic defects in CD3G lead to impaired expression of CD3 and TCR molecules on the cell surface and overall defective T-cell response to mitogenic signals, which was especially profound upon stimulation with PHA. Furthermore, impaired cell division was also observed for CD8+ (but not CD4+) T cells from CD3γ-deficient patients upon in vitro activation with anti-CD3. Similarly, a more profound impairment of T-cell proliferation in a mixed leukocyte reaction had been previously reported for CD8+ than for CD4+ T cells from CD3γ-deficient patients.16
Treg cells are dependent on TCR signaling for their development and suppressive function. In this study, we have shown that defects of TCR/CD3 expression and signaling in patients with CD3G mutations are associated with a reduced proportion of Treg cells, restriction of Treg repertoire diversity, and impaired Treg function. Previous studies have shown that diversity of TCR repertoire is maximal in naïve T cells, lower in T memory, and lowest in TEMRA cells,37,38 and that among memory T cells, richness of T-cell repertoire is higher in CD4+ than in CD8+ T cells.39 We observed a trend for reduced TRB repertoire diversity among patient-derived CD8+ T cells. This may reflect the accumulation of effector memory and TEMRA CD8+ cells in the patients. Although the reduced number of naïve T cells and the slightly reduced diversity of CD8+ T cells may play contribute to increased susceptibility to infections in CD3γ deficiency, a significant restriction of repertoire diversity was observed only in the Treg compartment, and was more prominent in patients P1 and P4. Interestingly, these were also the patients with the most severe autoimmune manifestations. Overall, these data strongly indicate that abnormalities of Treg diversity play an important role in determining the severity of immune dysregulation in CD3γ-deficient patients, and suggest that similar abnormalities may be at play also in other forms of PID with defective TCR signaling.
Analysis of amino acid composition at positions 6 and 7 of the TRB-CDR3 disclosed a molecular signature indicative of increased self-reactivity in CD4+ Tconv cells. TCR affinity for peptides and TCR signaling strength dictate the fate of developing CD4+ T cells in the thymus medulla. In particular, single positive CD4+ cells that recognize with high affinity the self-peptides that are presented by medullary thymic epithelial cells or by dendritic cells, undergo negative selection. Alternatively, self-reactive T cells may be diverted to natural Treg cells.40 This determines important differences in the antigen specificity of Treg vs CD4+ Tconv cells, with the former being enriched for self-reactive specificities.14 Abnormalities of TCR signaling strength, and in particular reduced signaling as in patients with CD3γ deficiency, may affect this process, leading to an increase in self-reactive clones within the CD4+ Tconv population. Our results showing enrichment for amino acids that promote self-reactivity, and reduced usage of those limiting self-reactivity at positions 6 and 7 of the TRB-CDR3 in CD4+ Tconv cells from CD3γ-deficient patients are consistent with this hypothesis. We have recently documented similar abnormalities of TRB composition in CD4+ Tconv cells from patients with hypomorphic RAG mutations,41 another condition that is characterized by increased occurrence of autoimmune manifestations. In that case, however, it is likely that the altered fate of self-reactive CD4+ thymocytes are secondary to reduced expression of AIRE and of AIRE-dependent tissue-restricted peptides in the thymic medulla, but the ultimate effect would be similar, with an excess of self-reactive specificities in CD4+ Tconv cells that are exported to the periphery. Interestingly, no molecular signature of enhanced self-reactivity was found among CD8+ T cells. In this regard, the molecular mechanisms controlling T-cell tolerance in thymus medulla (including generation of Treg cells from CD25+ FOXP3− or CD25− FOXP3+ progenitor cells) are better defined for CD4+ cells, but remain largely unknown for CD8+ cells.42
Finally, although analysis of the Treg TRB-CDR3 did not reveal significant changes in overall self-reactivity in CD3γ-deficient patients compared with controls, reduced signaling via the TCR/CD3 complex may have important consequences for the survival and function of Treg cells in the periphery. Indeed, the relative amount of TCR signaling is essential to maintaining survival and function of Treg cells.19,20 We have demonstrated that Treg cells from CD3γ-deficient patients have defective suppressive activity. Whether their survival is also affected, and whether this may contribute to the marked restriction of TRB repertoire in this subset, requires further investigation.
In summary, our data suggest a model whereby human CD3γ deficiency impairs surface expression of CD3 and TCR molecules, reducing TCR signaling strength and thereby affecting T-cell development, selection, and fate. Ultimately, this results in enrichment for self-reactive specificities within CD4+ Tconv cells, which may undergo peripheral activation and expansion because of the reduced number and impaired function of Treg cells. Altogether, these abnormalities create an environment that promotes the development of autoimmunity in this condition.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Authorship
Contribution: J.H.R. and O.M.D. performed the experiments, analyzed the data, and wrote the manuscript; B.D.S. and E.S.H. performed analysis of self-reactivity index; A.K.D., S.C.C., and Y.Y. performed experiments; L.A.H. contributed to the analysis of TRB repertoire; S.K., L.M.A., F.A.B., L.I.G.-G., S.N.G., H.K., C.Y., S.-Y.P., I.R., and J.R.R. provided patient samples and clinical information; and L.D.N. conceived the study, supervised all experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luigi D. Notarangelo, Laboratory of Clinical Immunology and Microbiology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Building 10, Room 5-3950, 10 Centre Dr, Bethesda, MD 20892; e-mail: luigi.notarangelo2@nih.gov.
References
Author notes
J.H.R. and O.M.D. contributed equally to this study.
I.R., J.R.R., and L.D.N. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal