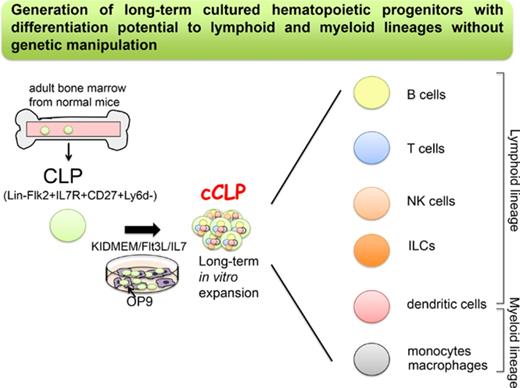
Key Points
We have established a novel culture system for long-term proliferating murine lymphoid progenitors without any genetic manipulation.
The cultured lymphoid progenitors can differentiate to lymphoid and myeloid lineages in vitro and in vivo.
Abstract
Common lymphoid progenitors (CLPs) differentiate to T and B lymphocytes, dendritic cells, natural killer cells, and innate lymphoid cells. Here, we describe culture conditions that, for the first time, allow the establishment of lymphoid-restricted, but uncommitted, long-term proliferating CLP cell lines and clones from a small pool of these cells from normal mouse bone marrow, without any genetic manipulation. Cells from more than half of the cultured CLP clones could be induced to differentiate to T, B, natural killer, dendritic, and myeloid cells in vitro. Cultured, transplanted CLPs transiently populate the host and differentiate to all lymphoid subsets, and to myeloid cells in vivo. This simple method to obtain robust numbers of cultured noncommitted CLPs will allow studies of cell-intrinsic and environmentally controlled lymphoid differentiation programs. If this method can be applied to human CLPs, it will provide new opportunities for cell therapy of patients in need of myeloid-lymphoid reconstitution.
Introduction
T and B lymphocytes, dendritic cells (DC), natural killer (NK) cells, and recently identified innate lymphoid cells (ILCs) all originate from a type of cell, the common lymphoid progenitors (CLPs).1-6 The small number of CLPs (0.03% of bone marrow hematopoietic cells) develop from long-term repopulating hematopoietic stem cells via multipotent progenitors and lymphoid-primed multipotent progenitors (LMPPs).7 LMPPs and CLPs have lost megakaryocyte-erythrocyte potential and are uncommitted intermediates with plasticity and flexibility to develop to all subsets of differentiated lymphoid cells in vivo as well as in vitro.
Hematopoietic progenitor cell lines with lymphoid and myeloid lineage differentiation potential in vivo have been successfully generated in vitro by transgenic overexpression of genes encoding Hoxb88 or Id3,9 or by genetic loss of Pax5,10 Ebf1,11 or E2A12 that all lead to developmental arrest of the cells resulting in sustained proliferation in an appropriate culture condition. Although CLPs from normal mouse bone marrow have never been maintained as proliferating cells with stable differentiation potential,6,13,14 our previous results with Pax5−/− progenitors and our experience with serum-free cultures of pre-BI cells from fetal liver and bone marrow encouraged us to attempt long-term culturing of CLPs under serum-free conditions.
Here, we describe methods that, for the first time, allow the long-term proliferation and cloning of stably uncommitted CLPs in vitro of normal mice without any genetic modifications. The cultured CLPs, which we named “cCLPs,” require the combined stimulation by Flt3L and low doses of interleukin-7 (IL-7), in the presence of CXCL12-producing stromal cells for long-term maintenance and proliferation in serum-free medium. These progenitors can be efficiently transduced with retroviral vectors. Moreover, cCLPs are clonable with high efficiencies, allowing an easy isolation of single transduced cCLPs. Clones of cCLPs retain the potential to differentiate in vitro with high (1 in 2) cloning efficiencies to T and B cells, and to myeloid cells, but not to erythroid and megakaryocytic lineage cells. When transplanted into sublethally irradiated recipient mice, these cCLP lines and clones differentiate to mature lymphoid, and subsets of myeloid cell lineages.
CLPs play essential roles in health and disease of the hematopoietic cell system. Our cCLP lines and clones can be expanded to large quantities. This opens new opportunities for large-scale genomic, proteomic, metabolomics, and physiological analyses of intracellular signaling and gene expression programs during lymphoid cell commitment, and of environmental influences on these programs. Thus, our cCLPs should be ideal targets for molecular, genetic, and cellular studies not only of normal lymphoid differentiation from early, uncommitted CLPs to later, lineage-committed lymphoid cells but also of deregulated states of differentiation in immune-deficiencies, autoimmune diseases, and lymphoid tumor development.
Materials and methods
Preparation of culture medium
Culture medium was prepared in Iscove modified Dulbecco medium (Gibco) supplemented with 3.02 g/L of NaHCO3 (Sigma)/50 μM β-mercaptoethanol (Gibco)/0.03% primatone (Quest International)/1× kanamycin (Gibco)/1× nonessential amino acid (Gibco) together with 2% fetal bovine serum (FBS; Sigma) as conventional serum-containing medium (CM) or 1 mg/mL of delipified bovine serum albumin (Sigma), 10 μg/mL of human transferrin (Sigma), and 0.005% fatty-acid supplement (Sigma), which we designated as Kawano's modified–Iscove's modified–Dulbecco's modified–Eagle's minimum essential medium (KIDMEM).
Cell culture
Lin−IL7R+Flk2+CD27+Ly6d− CLP population was isolated from bone marrow of 6- to 8-week-old mice, and 1 × 103 cells were cultured in 0.2 mL of CM or KIDMEM with 25 ng/mL of Flt3L and different concentrations of IL-7 (0.1 or 1 ng/mL) in 96-well flat-bottom plates precoated with OP9 stromal cell lines. Cultured cells were passaged on every 3 or 4 days to another well. Full details are described in the supplemental Methods, available on the Blood Web site.
In vitro cell differentiation
Lin−IL7R+Flk2+CD27+Ly6d− cells purified from cCLPs cultured for 2 months were cultured in the appropriate conditions for B, T, NK, DC, and myeloid cell differentiation.
Details for all other procedures are described in the supplemental Methods.
Results
Establishment of genetically unmodified cell lines from CLP of bone marrow in serum-free cultures
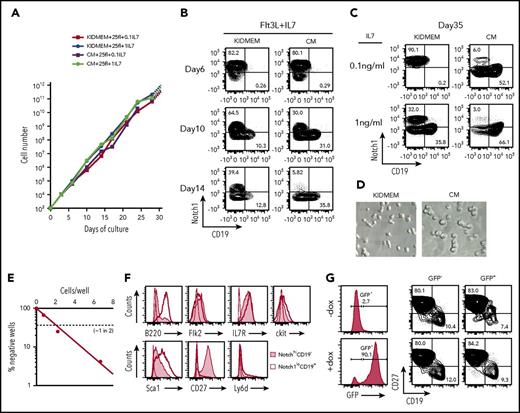
Uncommitted CLPs were fluorescence activated cell sorting (FACS)–enriched from bone marrow of C57BL/6 mice as Lin−IL7R+Flk2+CD27+Ly6d− cells15 and cultured on OP9 cells either in our modified serum-free medium, designated as KIDMEM, or in serum-containing CM, in the presence of Flt3L (25 ng/mL) and IL-7 at either 0.1 or 1 ng/mL. We examined Notch1 and CD19 expression on the cells after culture as markers for simple discrimination of CLP and precursor B cells as Notch1hiCD19− and Notch1loCD19+, respectively. Within 10 days, the number of cells increased between 1 and 3 × 103-fold, irrespective of KIDMEM or CM culture conditions and different concentrations of IL-7 (Figure 1A; supplemental Figure 1A). However, within the proliferating cells, the proportion of Notch1hiCD19− cells declined more rapidly in the CM than in the KIDMEM culture conditions, whereas Notch1loCD19− intermediates and Notch1loCD19+ B cells proportionally increased within 2 weeks (Figure 1B-C; supplemental Figure 1B-C,E). These findings reconfirm previous findings by other laboratories,6,14 that uncommitted CLPs cannot be maintained in CM cultures. By contrast, 40% of the cells cultured for 2 weeks in KIDMEM remained Notch1hiCD19− cells (Figure 1B). For the next 3 weeks, these cells preferentially expanded in KIDMEM with 0.1 ng/mL IL-7 so that >70% of the cells remained Notch1hiCD19− (Figure 1C; supplemental Figure 1D). This preferential expansion of these cultured Notch1hiCD19− cells continued for at least 2 more months at the same rate of proliferation (data not shown). The cultured cells displayed an extended shape (Figure 1D). Limiting dilution of purified Notch1hiCD19− cells from 2-month culture and further culture for 7 days in the same culture condition showed that ∼1 of 2 cells was clonally expandable (Figure 1E). The majority of clones and of the cells expanded in the clones retained the surface phenotype (supplemental Table 1). The concentration of IL-7 in cultures of CLP influenced their stability as uncommitted CLP. Thus, at 0.1 ng/mL IL-7, minor subpopulations of more differentiated Notch1loCD19− cells and Notch1loCD19+ pre-BI cells expanded with Notch1hiCD19− cells, and their proportion in the culture increased at 1 ng/mL IL-7 (Figure 1C; supplemental Figure 1C,E). Notchhi CD19− cells cultured for 1 month were FACS analyzed as Lin−IL7R+Flk2lockitlo/−Sca1loCD27+Ly6d−, consistent with the phenotypic markers for uncommitted CLPs from bone marrow, as previously described6,15 (Figure 1F). We call these cells, cultured from ex vivo CLP, “cCLP.”
Long-term proliferation of CLPs in vitro, and retroviral transduction of the cCLPs. FACS-purified Lin−Flk2+IL7R+CD27+Ly6d− cells (1 × 103) were cultured on irradiated OP9 stromal cells in our modified serum-free medium (KIDMEM) or serum-containing CM with Flt3L (25 ng/mL) and different concentrations of IL-7 (0.1 and 1 ng/mL). Cells at the indicated time points were counted and FACS analyzed for CD19 and Notch1 expression. (A) Kinetics of cell proliferation during long-term culture with high (circles) or low (squares) amount of IL-7 in KIDMEM or CM. (B-C) Representative FACS profiles of cells cultured with Flt3L and 0.1 ng/mL IL-7 at early phase up to day 14 (B) or cultured with Flt3L and different concentrations of IL-7 at day 35 (C) in KIDMEM or CM. The values in each panel indicate percentages of Notch1hiCD19− and Notch1loCD19+ cells. (D) Light microscopic images (20× objective) of cells at day 35 of culture with 0.1 ng/mL IL-7 in KIDMEM (left panel) or CM (right panel). (E) Limiting dilution of purified CLP cultured for 2 months under CLP conditions. (F) FACS analyses of Notch1hiCD19− CLP (shaded areas) or of Notch1loCD19+ B-lineage cells (thick lines) at day 35 of culture. Isotype-matched control is shown in thin lines. (G) Retroviral transduction of cCLPs with a vector containing EGFP under the expression controls of doxycycline. Representative FACS profiles of green fluorescent protein (GFP) and CD27/CD19 expression with (bottom) or without doxycycline (top) are shown.
Long-term proliferation of CLPs in vitro, and retroviral transduction of the cCLPs. FACS-purified Lin−Flk2+IL7R+CD27+Ly6d− cells (1 × 103) were cultured on irradiated OP9 stromal cells in our modified serum-free medium (KIDMEM) or serum-containing CM with Flt3L (25 ng/mL) and different concentrations of IL-7 (0.1 and 1 ng/mL). Cells at the indicated time points were counted and FACS analyzed for CD19 and Notch1 expression. (A) Kinetics of cell proliferation during long-term culture with high (circles) or low (squares) amount of IL-7 in KIDMEM or CM. (B-C) Representative FACS profiles of cells cultured with Flt3L and 0.1 ng/mL IL-7 at early phase up to day 14 (B) or cultured with Flt3L and different concentrations of IL-7 at day 35 (C) in KIDMEM or CM. The values in each panel indicate percentages of Notch1hiCD19− and Notch1loCD19+ cells. (D) Light microscopic images (20× objective) of cells at day 35 of culture with 0.1 ng/mL IL-7 in KIDMEM (left panel) or CM (right panel). (E) Limiting dilution of purified CLP cultured for 2 months under CLP conditions. (F) FACS analyses of Notch1hiCD19− CLP (shaded areas) or of Notch1loCD19+ B-lineage cells (thick lines) at day 35 of culture. Isotype-matched control is shown in thin lines. (G) Retroviral transduction of cCLPs with a vector containing EGFP under the expression controls of doxycycline. Representative FACS profiles of green fluorescent protein (GFP) and CD27/CD19 expression with (bottom) or without doxycycline (top) are shown.
Retroviral transduction of cCLP
cCLPs cultured for 2 months were transduced with a doxycycline-inducible retroviral vector containing a gene encoding enhanced green fluorescent protein (EGFP). Within 4 days, EGFP-expressing cCLPs were expanding by proliferation in the presence of doxycycline (Figure 1G). Thus, genes of choice can be transduced in cCLP by retroviral vectors.
Molecular control of proliferation and differentiation of cCLP
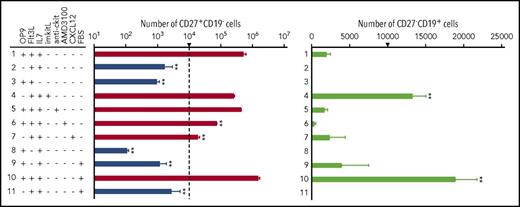
To evaluate the roles of stromal cells and of the cytokines Flt3L and IL-7 in the long-term maintenance and proliferation of cCLPs, we replated Lin−-IL7R+Flk2loCD27+Ly6d− cells purified from 1-month culture and measured their state of differentiation, survival, and proliferative capacity after 7 days under different culture conditions. In the presence of OP9 stromal cells, Flt3L, and 0.1 ng/mL IL-7, cCLPs (CD27+CD19−) proliferated and retained their undifferentiated state (Figure 2 first row; supplemental Figure 2A-B). When IL-7 or the OP9 stromal cells were omitted from culture, cCLPs did not continue to proliferate (Figure 2 second and third row). Plate-immobilized recombinant kit-ligand, which we previously found to be able to replace OP9 stromal cells in pre–BI-cell cultures,16 could not replace OP9 stromal cells to maintain the CLP phenotype in KIDMEM cultures with 0.1 ng/mL IL-7, but rather promoted pre–BI-cell (CD27−CD19+) proliferation and differentiation (Figure 2 fourth row). Antibodies against c-kit, which had previously been found to fully inhibit proliferation of the more differentiated pre–BI cells in stromal cell/IL-7 cultures,17 had only a minor inhibitory effect on cCLP proliferation (Figure 2 fifth row). On the other hand, AMD3100, the CXCR4-specific inhibitor of CXCL12-mediated signaling, inhibited cCLP proliferation at least 10-fold, without affecting their survival and phenotype (Figure 2 sixth row). These results suggest that IL-7/IL-7R-mediated signaling as well as CXCL12/CXCR4 signaling, but not kitL/c-kit signaling, have major influences on the proliferation of stably uncommitted cCLPs, and that stromal cells appear to supply CXCL12 in our cultures. However, we found that the addition of soluble, recombinant CXCL12 did not abolish the requirement for OP9 stromal cells (Figure 2 seventh row), suggesting that additional properties of stromal cells are contributing to AMD3100-inhibitable cCLP proliferation in vitro.
Ligand-controlled proliferation, survival, and differentiation of cCLPs. FACS-purified Lin−Flk2+IL7R+CD27+Ly6d− cells from 1-month cCLPs were further cultured for another 7 days, starting from 1 × 104 cells on 24-well flat-bottom plates at day 0 (this number of cells at the start of the cultures is indicated by an arrow), in various combinations with OP9, Flt3L (25 ng/mL), IL-7 (0.1 ng/mL), immobilized kitL (imkitL, 1 μg/mL), CXCL12 (100 ng/mL), FBS (2%), anti-ckit monoclonal antibody (10 μg/mL), and/or AMD3100 (10 μM). Statistical analysis of numbers of CD27+CD19− (logarithmic scale, left panel) and CD27−CD19+ cells (linear scale, right panel) are shown. On the left panel, cell numbers in culture at day 7 <104 (ie, decreased numbers within these 7 days), are drawn in blue whereas those >104 are drawn in red, indicating increased numbers within 7 days. Error bars indicate mean (n = 3) ± standard deviation. **P < .01. These data are representative of 3 independent experiments.
Ligand-controlled proliferation, survival, and differentiation of cCLPs. FACS-purified Lin−Flk2+IL7R+CD27+Ly6d− cells from 1-month cCLPs were further cultured for another 7 days, starting from 1 × 104 cells on 24-well flat-bottom plates at day 0 (this number of cells at the start of the cultures is indicated by an arrow), in various combinations with OP9, Flt3L (25 ng/mL), IL-7 (0.1 ng/mL), immobilized kitL (imkitL, 1 μg/mL), CXCL12 (100 ng/mL), FBS (2%), anti-ckit monoclonal antibody (10 μg/mL), and/or AMD3100 (10 μM). Statistical analysis of numbers of CD27+CD19− (logarithmic scale, left panel) and CD27−CD19+ cells (linear scale, right panel) are shown. On the left panel, cell numbers in culture at day 7 <104 (ie, decreased numbers within these 7 days), are drawn in blue whereas those >104 are drawn in red, indicating increased numbers within 7 days. Error bars indicate mean (n = 3) ± standard deviation. **P < .01. These data are representative of 3 independent experiments.
When Flt3L was omitted from the serum-free KIDMEM cultures, cCLPs did not proliferate and entered apoptosis. The few surviving cells differentiated to CD27−CD19+ pre–BI cells (Figure 2 eighth row). Addition of FBS enhanced this differentiation (Figure 2 ninth row) and further supported the proliferation of both cCLPs and pre–BI cells in the presence of OP9 stromal cells, Flt3L, and IL-7 (Figure 2 10th row). Again, removal of OP9 from this culture condition abolished cCLP proliferation and pre–BI-cell differentiation (Figure 2 11th row). This could also be monitored by the upregulation of B-cell–specific genes such as Ebf1, Pax5, and Igll1 (supplemental Figure 2C). From the differentiated CD19+ cells, long-term proliferating pre–BI-cell lines could be established on stromal cells in the presence of high concentrations of IL-7 (supplemental Figure 3).
From all these results, it is apparent that Flt3L-Flt3 interactions stabilize the uncommitted state of cCLP differentiation and allow their survival, and that FBS can overcome this stabilizing influence.
High similarities of gene expression programs of cCLP and CLP ex vivo by whole transcriptome analysis
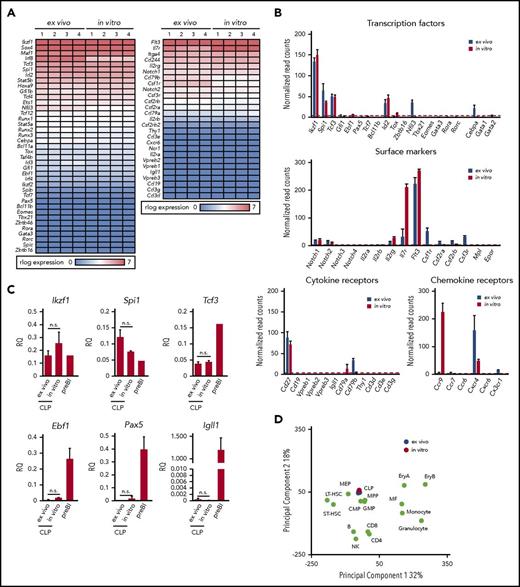
In order to see how similar cCLPs cultured for 2 months are in comparison with CLPs isolated ex vivo from bone marrow (ie, with the original cells put in culture), we performed transcriptome analyses by RNA-sequencing (RNA-seq). It can be seen from a representative set of genes shown in Figure 3A and from the data deposited (GSE109805), that the gene expression patterns were remarkably similar. Principal component analysis of our RNA-seq data from cCLP and CLP ex vivo also supported a higher similarity between them, compared with other hematopoietic lineages (Figure 3D). As expected from the FACS-determined surface protein phenotype of these cells (Figure 1E), this messenger RNA expression analysis detected Notch1, Cd27, Cxcr4, Flt3, and Il7ra on both of them, although Il7ra expression was significantly higher in cCLP than in CLP ex vivo (Figure 3B). cCLP expressed transcription factors that have been previously reported to function in CLP, such as Tcf3 (E2A),18,19 a critical factor for the generation of LMPP and CLP, Ikzf1(Ikaros),20 required for Flt3 expression and Spi1 (PU.1)21 at almost the same levels as in CLP ex vivo (Figure 3B). These results were validated by quantitative reverse transcription polymerase chain reaction analyses (Figure 3C). Impressively, both of these cells showed low or undetectable levels of messenger RNAs expression encoding transcription factors involved in lymphoid lineage specification, such as Ebf1 and Pax5 for B cells, Tcf7, Bcl11b for T cells, Eomes for NK cells, Tox, Tbx21, Gata3, Rora, and Rorc, Zbtb16 for ILCs, although Id2 and Nfil3 were similarly or differentially expressed in both cCLP and CLP ex vivo (Figure 3B). Quantitative reverse transcription polymerase chain reaction analyses validated a much lower expression of Ebf1, Pax5, and Igll1 in both of them, as compared with long-term cultured CD19+ pre–BI cells (Figure 3C). Genes linked to hematopoietic stem cells, such as Tal1, Runx1, Hoxb5, and Fdg5, were expressed in neither of them (data not shown). Moreover, both cCLPs and CLPs ex vivo expressed very low or undetectable levels of genes encoding signaling components of antigen receptors for B cells or T cells, such as mb-1 (Igα), B29 (Igβ), or CD3g, CD3d, CD3e, respectively, as representative genes of their precursors such as VpreB, Igll1, or Ptcra (pre–T-cell receptor α) (Figure 3B, see also deposited data). Genes associated with myeloid-lineage specification, including Cebpa, Csf1r (M-CSFR), Csf2ra, Csf2rb (GM-CSFR), and Csf3r (for G-CSFR), were also expressed only at low or undetectable levels, even lower in cCLPs than in CLP ex vivo (Figure 3B). These results suggest that cCLPs display gene expression profiles for surface molecules, transcription factors, and signaling molecules involved in lymphoid differentiation potential in vivo that closely resemble those of CLP ex vivo.
Gene expression profiles of cCLPs and CLPs ex vivo. (A) Heat map view of representative gene expression profiles on transcription factors and receptors in cCLPs (in vitro) and CLPs ex vivo (n = 4). (B) Statistical analyses of representative gene expression of transcription factors (top), surface markers (middle), Notch family and cytokine receptors (lower left), and chemokine receptors (lower right) associated with the hematopoietic cell development measured by RNA-seq in cCLP (red bars) or CLP ex vivo (blue bars). Read counts after normalization were used as gene expression (see “Materials and methods”). Error bars indicate mean (n = 4) ± standard deviation. (C) Quantitative reverse transcription polymerase chain reaction analysis of CLP- or B-cell–related gene expression (n = 4). Relative quantities (RQ) of the indicated genes normalized to the expression of Gapdh are shown. (D) Two-dimensional principal component analysis of normalized whole gene expression profiles in cCLPs (in vitro, n = 4, in red), freshly isolated CLPs (ex vivo, n = 4, in blue), and other hematopoietic cells (in green). preBI, precursor BI (B220+CD19+ckit+IL7R+DHJH/DhJH-rearranged). n.s., not significant.
Gene expression profiles of cCLPs and CLPs ex vivo. (A) Heat map view of representative gene expression profiles on transcription factors and receptors in cCLPs (in vitro) and CLPs ex vivo (n = 4). (B) Statistical analyses of representative gene expression of transcription factors (top), surface markers (middle), Notch family and cytokine receptors (lower left), and chemokine receptors (lower right) associated with the hematopoietic cell development measured by RNA-seq in cCLP (red bars) or CLP ex vivo (blue bars). Read counts after normalization were used as gene expression (see “Materials and methods”). Error bars indicate mean (n = 4) ± standard deviation. (C) Quantitative reverse transcription polymerase chain reaction analysis of CLP- or B-cell–related gene expression (n = 4). Relative quantities (RQ) of the indicated genes normalized to the expression of Gapdh are shown. (D) Two-dimensional principal component analysis of normalized whole gene expression profiles in cCLPs (in vitro, n = 4, in red), freshly isolated CLPs (ex vivo, n = 4, in blue), and other hematopoietic cells (in green). preBI, precursor BI (B220+CD19+ckit+IL7R+DHJH/DhJH-rearranged). n.s., not significant.
Among genes upregulated in cCLPs, but not in CLPs ex vivo, was Ccr9, encoding a chemokine receptor active in lymphoid progenitor cell homing to the thymus (Figure 3B).22-24 Thus, the long-term proliferating cCLP could assume the phenotype of a subset of CLPs at an intermediate stage on its way to become what has previously been defined as CD4+/−CD25−CD44+c-kit+CD27+ Flt3+CCR9+ thymus-settling cells.25
cCLPs differentiate to lymphoid and myeloid cell lineages in vitro
Next, we examined the differentiation potential of cCLPs, expanded in culture for 2 months. We induced differentiation of cCLPs to CD19+ B-lineage cells in CM on OP9 stromal cells in the presence of IL-7 (1 ng/mL) (Figure 4A upper left). T-cell differentiation can be induced by culturing CLP on delta-like 1-expressing OP9 (OP9DL1) stromal cells in the presence of Flt3L and low doses of IL-7.26 Under the T-cell–differentiating conditions for 7 days, cCLPs developed to CD44+CD25− CD4−CD8− double-negative thymocyte 1 (DN1), CD44+CD25+ DN2, and a few CD44−CD25+ DN3 thymocytes (Figure 4A upper middle). DC development involves several transcription factors, including Ikaros and PU.1, and the orchestrated signaling through Flt3L, macrophage colony-stimulating factor (M-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF).5 We differentiated cCLPs in the presence of Flt3L, IL-6, M-CSF, and GM-CSF. Within 14 days, CD11c+MHCII+ DCs with typical dendritic morphology developed (Figure 4A middle panel). Because they were also CD11b+ F4/80+ Ly6C−Ly6G−, they appear to be classical DCs, although this phenotypic analysis cannot exclude the possibility, that they are monocyte-derived DC (supplemental Figure 4C). These DCs were capable of presenting antigen to stimulate T-cell proliferation (supplemental Figure 4A). Furthermore, a mixture of the cytokines Flt3L, SCF, IL3, IL-6, IL-7, M-CSF, GM-CSF, and TNFα induced cCLPs to CD11b+Gr1+ myeloid-lineage cells (Figure 4A lower left). These cells included F4/80+Ly6c+CD11c−Ly6G−MHCII−FSCloSSClo and F4/80+Ly6c−CD11cloLy6GloMHCII−FSChiSSChi populations, corresponding to monocytes and macrophages, respectively (Figure 4A lower middle; supplemental Figure 4C). Giemsa staining of these myeloid cells showed extended cytoplasm with azurophilic granules, characteristic of macrophages (Figure 4A lower right). Gr1hi or Ly6Ghi neutrophils were not observed in the culture condition (data not shown). These myeloid lineage cells, but not the uncommitted cCLPs, were capable of typical macrophage-associated phagocytosis of fluorescent Escherichia coli (supplemental Figure 4B).
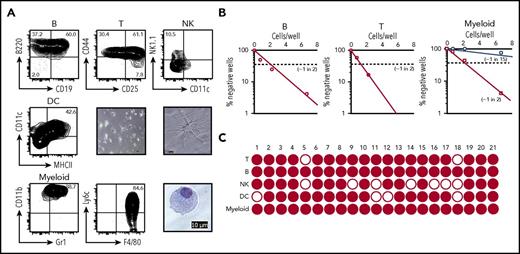
In vitro differentiation of cCLP into lineage-positive cells. (A) Representative FACS profiles on differentiation to B cells (upper left), T cells (upper middle), NK cells (upper right), and myeloid cells (lower left and middle) at day 7, and to DC (middle left) at day 14 in different culture conditions of Lin−Flk2+IL7R+CD27+Ly6d− cells FACS-purified from cCLP cultured for 2 months. Microscopic images of myeloid cells (lower right) and DC differentiated from cCLP with low (center panel) and high (middle right) magnification. Scale bar, 10 μm. (B) Limiting dilution analyses of 2-month cCLPs for frequencies of proliferating clones under B-cell (left), T-cell (middle), or myeloid cell differentiation conditions (right). For myeloid cell differentiation, cCLPs were cultured on irradiated OP9 (circles) or nonirradiated OP9 (squares). (C) Single-clone analysis of cCLP for the differentiation to B (CD19+), T (Thy1+), NK (CD122+), DC (CD11c+MHCII+), and myeloid cells (CD11b+Gr1+). Wells that included >10 lineage-positive cells counted by flow cytometer were indicated as closed circle. For differentiation conditions, see “Materials and methods.” All data are representative of 2 independent experiments.
In vitro differentiation of cCLP into lineage-positive cells. (A) Representative FACS profiles on differentiation to B cells (upper left), T cells (upper middle), NK cells (upper right), and myeloid cells (lower left and middle) at day 7, and to DC (middle left) at day 14 in different culture conditions of Lin−Flk2+IL7R+CD27+Ly6d− cells FACS-purified from cCLP cultured for 2 months. Microscopic images of myeloid cells (lower right) and DC differentiated from cCLP with low (center panel) and high (middle right) magnification. Scale bar, 10 μm. (B) Limiting dilution analyses of 2-month cCLPs for frequencies of proliferating clones under B-cell (left), T-cell (middle), or myeloid cell differentiation conditions (right). For myeloid cell differentiation, cCLPs were cultured on irradiated OP9 (circles) or nonirradiated OP9 (squares). (C) Single-clone analysis of cCLP for the differentiation to B (CD19+), T (Thy1+), NK (CD122+), DC (CD11c+MHCII+), and myeloid cells (CD11b+Gr1+). Wells that included >10 lineage-positive cells counted by flow cytometer were indicated as closed circle. For differentiation conditions, see “Materials and methods.” All data are representative of 2 independent experiments.
We attempted NK and ILC differentiation from cCLP. In the presence of IL-2 and IL-15, cCLPs differentiated to NK1.1+NK cells (Figure 4A upper right), but these cells could not kill YAC1 targets (data not shown). Also, more differentiated ILCs remained undetectable. cCLPs could also not be induced to differentiate to Ter119+ or CD41+ erythroid-lineage cells (data not shown). These results suggest that these cCLPs retain a lymphoid-uncommitted status, allowing them to differentiate in vitro not only to the major lymphoid lineages but also to myeloid lineages.
Differentiation of cCLP clones
Because cCLPs can be cloned, we tested single cCLP clones for their lymphoid differentiation potential in culture conditions, which promote proliferation, but also favor differentiation into different myeloid and lymphoid lineages (see “Materials and methods”). Limiting dilution assays of 2-month cultured cCLP clones, maintained for 7 days under B-lymphoid differentiation conditions, showed that ∼1 of 2 clones developed to CD19+ B-lineage cells (Figure 4B left). Again, ∼1 in 2 clones differentiated to Thy1+T-lymphoid cells (Figure 4B middle). These results indicate that single cCLPs have the differentiation potential to B- and T-lymphoid lineage cells in vitro. Interestingly, differentiation to myeloid cells depended on the status of the OP9 stromal cells. On irradiated OP9, 1 in 15 clones differentiated, whereas ∼1 in 2 cells did so on nonirradiated OP9 (ie, with frequencies similarly as high as those for T- and B-lineage cells) (Figure 4B right). All of the 21 cCLP clones expanded from 2-month cultured cCLPs could be induced to differentiate to myeloid and B lineages, 19 of them also to T lineages, 14 of them to NK lineages, and 17 of them to DC lineages. Twelve of the 21 clones could differentiate to all tested lineages (Figure 4C; supplemental Table 2). Similar frequencies of clones with multiple B-, T-lymphoid, and myeloid differentiation potential were observed with clones of CLP isolated ex vivo and freshly expanded (supplemental Figure 5).
These results suggest that at least a large part of cCLP clones retain differentiation potential to lymphoid and myeloid lineages in vitro.
In vivo differentiation potential of cCLPs
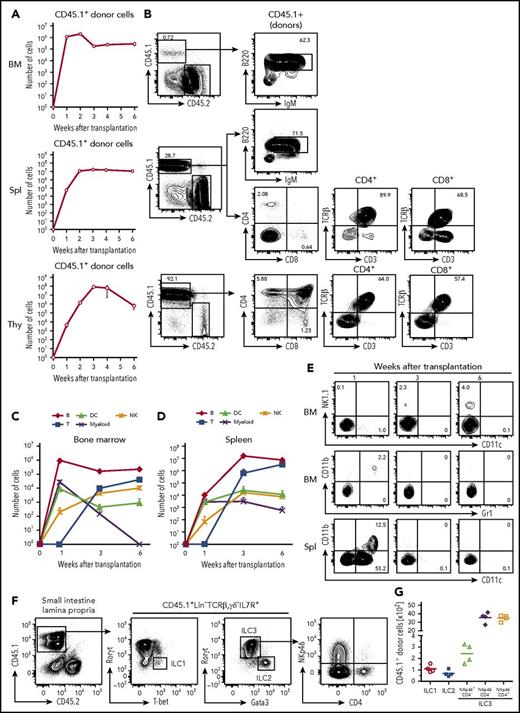
To examine the differentiation potential of cCLP in vivo, 1 × 106 CD45.1+ cCLPs cultured for 2 months were transplanted IV into sublethally irradiated congenic CD45.2+ mice. One and 2 weeks after transplantation, the majority of CD45.1+ donor cells were found in bone marrow and a smaller number were found in spleen (Figure 5A). Most of them had lost their CLP phenotype. At this early time after transplantation, the donor-derived cells were mostly B220+IgM− precursor B cells and B220+IgM+ immature B cells (Figure 5A-B).
Differentiation potentials of cCLPs in vivo. CD45.1+cCLP cultured for 2 months were transplanted into sublethally irradiated CD45.2+C57BL/6 mice (n = 4). Bone marrow (top), spleen (middle), and thymus (bottom) of the recipient hosts were FACS analyzed at the indicated times after transplantation. (A) Total numbers of CD45.1+ donor cells. (B) Representative FACS profiles of donor-derived T and B cells differentiated at 3 weeks after transplantation in bone marrow (upper panel), spleen (middle panel), and thymus (lower panel). (C-D) Total numbers of indicated cell subsets derived from donors in bone marrow (C) and spleen (D) at different times after transplantation. (E) Representative FACS profiles of donor-derived NK1.1+NK cells (upper panel), CD11b+Gr1+ myeloid cells (middle panel) in bone marrow, and CD11b+CD11c+, CD11b−CD11c+ DCs in spleen (lower panel) at different times after transplantation. The values in each panel shown in panels B and E indicate the percentages of cells in each gate. Data shown in panels A to E are representative of 2 independent experiments. (F) Representative FACS profiles and (G) total numbers of donor-derived ILC subsets in intestine 4 weeks after transplantation of CD45.1+ cCLPs cultured for 2 months into sublethally irradiated CD45.2+ RAG2/c-deficient mice (n = 4). BM, bone marrow; IgM, immunoglobulin M; Spl, spleen; Thy, thymus.
Differentiation potentials of cCLPs in vivo. CD45.1+cCLP cultured for 2 months were transplanted into sublethally irradiated CD45.2+C57BL/6 mice (n = 4). Bone marrow (top), spleen (middle), and thymus (bottom) of the recipient hosts were FACS analyzed at the indicated times after transplantation. (A) Total numbers of CD45.1+ donor cells. (B) Representative FACS profiles of donor-derived T and B cells differentiated at 3 weeks after transplantation in bone marrow (upper panel), spleen (middle panel), and thymus (lower panel). (C-D) Total numbers of indicated cell subsets derived from donors in bone marrow (C) and spleen (D) at different times after transplantation. (E) Representative FACS profiles of donor-derived NK1.1+NK cells (upper panel), CD11b+Gr1+ myeloid cells (middle panel) in bone marrow, and CD11b+CD11c+, CD11b−CD11c+ DCs in spleen (lower panel) at different times after transplantation. The values in each panel shown in panels B and E indicate the percentages of cells in each gate. Data shown in panels A to E are representative of 2 independent experiments. (F) Representative FACS profiles and (G) total numbers of donor-derived ILC subsets in intestine 4 weeks after transplantation of CD45.1+ cCLPs cultured for 2 months into sublethally irradiated CD45.2+ RAG2/c-deficient mice (n = 4). BM, bone marrow; IgM, immunoglobulin M; Spl, spleen; Thy, thymus.
At 1 week, but not later, small numbers of donor-derived CD11b+Gr1+ myeloid cells were detected in bone marrow, and donor-derived CD11b−CD11c+ or CD11b+CD11c+ DCs were detected in spleen (Figure 5C-E). Donor-derived mature granulocytes or erythroid-lineage cells could not be detected at any time up to 6 weeks after transplantation (data not shown). Three weeks after transplantation, the number of donor-derived B220+IgM− precursor B cells and B220+IgM+ immature B cells started to decline in bone marrow and spleen (Figure 5A-B). Later, between 3 and 4 weeks after transplantation, donor-derived CD4+CD8+ thymocytes appeared in the thymus, and CD4+ as well as CD8+ T cells, expressing T-cell receptor β and CD3, appeared in spleen (Figure 5A-B). Donor-derived NK1.1+ NK cells were found in bone marrow and spleen, increasing in numbers at later times after transplantation (Figure 5C-E). Competitive transfer analysis of cCLPs with CLPs ex vivo revealed an ∼1/200 lower repopulation potential, but a similar differentiation potential of cCLPs to those of CLPs ex vivo (supplemental Figure 6A-D). In vivo clonal analysis of 4 randomly selected single cCLP clones showed that all clones differentiated to B, T, and NK cells, but no myeloid cells were detectable (supplemental Figure 6E).
All 3 ILC populations, namely donor-derived T-bet+ ILC1, Gata3hi ILC2, and RORγt+ ILC3 subsets, as defined by NKp46 and CD4 expression were detected in the small intestinal lamina propria of Rag2γc-deficient mice 4 weeks after transplantation of cCLP (Figure 5F-G). Moreover, the main ILC populations residing in lung, in particular Eomes+ NK cells and Gata3hi ILC2, or in the liver, specifically T-bet+ ILC1 and Eomes+ NK cells, could be reconstituted (supplemental Figure 7). Comparable numbers of B, T, NK cells from donors were obtained in control experiments, where CLPs, isolated ex vivo from bone marrow, were transplanted (supplemental Figure 8). Transplantations of cCLPs into RAG2/γc-deficient hosts yielded lower numbers of T cells and B cells (supplemental Figure 9). Transplantations of cCLPs cultured for >4 months yielded lower numbers of donor cells, compared with those cultured for 2 months (supplemental Figure 10).
From all these transplantation experiments, we conclude that our long-term cultured, genetically unmodified cCLPs have the potential to differentiate in vivo to all lymphoid cell lineages tested.
Discussion
For the first time, we have been able to propagate CLPs from bone marrow of normal mice for weeks and months as long-term proliferating cell lines and clones, which closely resemble ex vivo CLP. CLP defined as Lin−ckitloSca1loIL7R+Thy1−6, later refined as LinloFlk2+IL7R+CD27+, were found to possess myeloid potential in vitro, but were found to display little myeloid potential in vivo.27 On the other hand, LMPP, defined as Lin−ckit+Sca1+Flk2+, were shown to have stronger myeloid potential in vitro and in vivo, and colonies with myeloid potential were detected in spleen and bone marrow a week after the in vivo transplantation.7 We have established the cell lines described here from Lin−Flk2+IL7R+CD27+Ly6d−CLP. Because we find them to have differentiation capacities for lymphoid and myeloid lineages in vitro and in vivo, we call these lines “cCLP (cultured CLP),” in order to distinguish them from ex vivo CLPs, or ex vivo LMPPs. Karyotype test by Q-band method of the 2-month cCLPs revealed that all of cells tested retained a full set of chromosomes (data not shown). It has been known that some hematopoietic cells, such as CD4+ and CD8+ T cells, CD19+ckit+IL7R+preBI cell lines, and FceRI+ckit+IL3R+bone marrow-derived mast cells, can be generated and propagated long term in vitro from primary lymphoid tissues without any genetic manipulation, but all of them are already lineage committed with reduced or specialized differentiation potential. Thus, this is the first report that describes the establishment of long-term expanding hematopoietic uncommitted progenitor lines and clones with differentiation potential in vitro and in vivo without any genetic manipulations or modifications. Our genetically unaltered cCLP lines and clones are a significant advance over previous studies, in which transgenic overexpression of Hoxb88 or Id3,9 or germline deletion of Pax5,10 Ebf1,11 or E2A,12 had led to developmental arrests of progenitors resembling multipotent progenitors or CLP, and had enabled their establishment as long-term proliferating, stably differentiated, but genetically altered cell lines and clones.
In order to achieve the same success with normal, genetically unaltered cells, we have introduced several changes in tissue culture conditions and introduced a modified version of Iscove modified Dulbecco medium, KIDMEM, in which FBS is replaced by purified transferrin and delipified bovine serum albumin, complexed with a defined mixture of lipids.28 This has removed hematopoietic and B-cell differentiation-inducing activities of serum.29 We have used the well-documented synergistic action of IL-7 and Flt3L,6,13,14 but we have lowered the concentration of IL-7 to 0.1 ng/mL, reducing its B-cell differentiation-inducing activity, to stimulate proliferation of CLP, a condition that we find to inhibit the induction of Ebf1 and Pax5 expression, hence, to prevent the commitment to B-cell differentiation. Other laboratories have found that Flt3L and IL-7 cooperate to improve B-cell development.13,14 However, in these previous experiments, FBS was present in the cultures, which we show here to have strong B-cell differentiation-inducing activities. Our finding, that Flt3L stabilizes the uncommitted CLP state of differentiation, is a novel view of the action of Flt3L during hematopoietic and B-lymphopoietic differentiation. This stabilizing action is a crucial element of the success of the long-term culturing of cCLP. OP9 stromal cells and low doses of IL-7 both contribute to proliferation of cCLPs, whereas kit-ligand is only marginally active. Therefore, the differentiation from CLP to pre–BI cells coincides with a major change in the controls of proliferation to a kitL/c-kit–controlled signaling. The CXCR4-inhibitor AMD3100 inhibits proliferation of cCLPs but allows undifferentiated cCLPs to survive. Thus, IL-7 and Flt3L are major candidates for ligands, which could signal cCLP to survive without further differentiation. Furthermore, the M-CSF deficiency of the OP9-stromal cells might avoid a possible, c-fms–mediated myeloid differentiation of the cCLPs, and this suggestion is supported by our findings that M-CSF–proficient stromal cells (eg, ST-2) do not support the long-term maintenance of cCLPs (data not shown). Neither soluble nor plate-immobilized recombinant CXCL12 could replace OP9 stromal cells in proliferating CLP cultures, suggesting a more complex form of growth support (Figure 2 and data not shown).
The comparison of the transcriptomes of cCLP with CLP ex vivo shows a high similarity between them. However, in contrast to ex vivo CLPs, cCLPs downregulate IL7Rα, Flt3, and CXCR4 surface expression, probably induced by the presence and action of their ligands in culture. On the other hand, these cell lines, but not CLP ex vivo, also express CD16 and CCR9, markers expressed on Lin−Flt3+IL7R−/loc-kithi early thymic precursors,25,30 that also retain B-cell and myeloid differentiation capacity.31,32 Expression of CCR9 on CLP facilitates their transfer to the thymus.33,34 Because Rag1 as well as Ccr9 expression was upregulated in cCLPs, they resemble hematopoietic progenitors known as Lin−Rag1+ earliest lymphoid progenitors (data deposited as GSE109805).35
Our method to obtain large numbers of hematopoietic progenitors with myeloid-lymphoid potential makes it possible to study the complex endogenous choices of molecular programs for hematopoietic lineage differentiation (ie, dynamic epigenetic regulations of genes involved in receptor-ligand interactions stimulating signaling pathways to gene expression controls), and influences of metabolic activities on CLP (eg, oxidative phosphorylation, glycolysis and lipolysis, autophagy, and hypoxia). It will also facilitate evaluations of influences of CLP by their environments. Our ability to express selected genes by retroviral transductions in our cCLPs predicts that better targeted introductions of genes into the genome (eg, by CRSPR-cas9 technology) will also be possible, opening exciting new possibilities to study lymphopoiesis from CLP.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE109805).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Hiromi Kubagawa and Koji Tokoyoda, both at DRFZ Berlin, for helpful discussions, and Hiromi Kubagawa and Simon Fillatreau for critical reading of the manuscript; Shintaro Hojyo and Peter Jani for helpful suggestions and discussions; and Kathrin Lehmann, Toralf Kaiser, and Jenny Kirsch for competent technical help. Y.K. thanks the Humboldt Foundation for support as a Humboldt Fellow.
This work was in part supported by a Kosellek Grant of the DFG (ME2764/1-1) (F.M.) and KAKENHI (Grant-in-Aid for Scientific Research).
Authorship
Contribution: Y.K., G.P., M.-F.M., C.R., H.K., and F.M. planned the experiments; Y.K., G.P., C.S., K.T., and G.A.H. performed the experiments; Y.K., C.S., and P.D. analyzed the data; and Y.K. and F.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yohei Kawano, Tokyo Medical and Dental University (TMDU), Department of Immune Regulation, Yushima 1-5-45, Bunkyo-ku, Tokyo 1138510, Japan; e-mail: youhei.mbch@tmd.ac.jp; and Fritz Melchers, Deutsches Rheuma-Forschungszentrum Berlin (DRFZ), Chariteplatz 1, 10117 Berlin, Germany; e-mail: fritz.melchers@unibas.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal