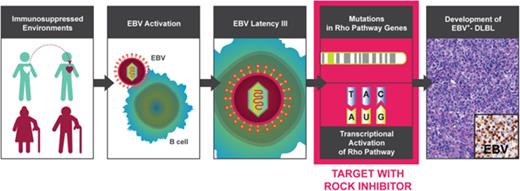
Key Points
EBV-induced DLBLs are characterized by genomic and transcriptomic alterations in the Rho pathway.
Targeting the Rho pathway using a ROCK inhibitor, fasudil, inhibited tumor growth in EBV-positive DLBL patient-derived xenograft models.
Abstract
Epstein-Barr virus (EBV)-positive diffuse large B-cell lymphomas (EBV+-DLBLs) tend to occur in immunocompromised patients, such as the elderly or those undergoing solid organ transplantation. The pathogenesis and genomic characteristics of EBV+-DLBLs are largely unknown because of the limited availability of human samples and lack of experimental animal models. We observed the development of 25 human EBV+-DLBLs during the engraftment of gastric adenocarcinomas into immunodeficient mice. An integrated genomic analysis of the human-derived EBV+-DLBLs revealed enrichment of mutations in Rho pathway genes, including RHPN2, and Rho pathway transcriptomic activation. Targeting the Rho pathway using a Rho-associated protein kinase (ROCK) inhibitor, fasudil, markedly decreased tumor growth in EBV+-DLBL patient-derived xenograft (PDX) models. Thus, alterations in the Rho pathway appear to contribute to EBV-induced lymphomagenesis in immunosuppressed environments.
Introduction
Epstein-Barr virus (EBV) is a major oncovirus causing several types of neoplasm in human, and the association between EBV and lymphomas is well documented.1 Diffuse large B-cell lymphoma (DLBL) associated with EBV has also been reported in EBV-positive DLBL (EBV+-DLBL) of the elderly, which was a newly defined lymphoma entity.2 EBV+-DLBL showed worse prognosis compared with EBV-negative DLBL.3,4
The development of EBV-induced lymphoma is associated with decreased immune function. Immune deficiency caused by HIV infection and solid organ transplantation increased the incidence of non-Hodgkin lymphoma (NHL) about 70-fold and 8-fold, respectively.5 Despite the introduction of highly active antiretroviral therapy, the decline in the incidence of EBV-related NHL was less marked than that of other HIV-associated malignancies, and NHL remains the most common HIV-associated malignancy.6 Most HIV-associated NHLs are B-cell neoplasms, including DLBL and Burkitt lymphoma,6 and DLBL occurs in more profound immune deficiency compared with Burkitt lymphoma.5 However, detailed pathogenesis and genomic characteristics of EBV-induced DLBL remain largely unknown because of limited sample availability from human.
Patient-derived xenografts (PDXs) are mouse tumor models generated by implanting patient tumor tissue fragments into immunodeficient mice.7 In PDX models of several cancer types, human-derived lymphomagenesis has been reported.8-12 These lymphomas were mostly of a B-cell subtype and highly associated with EBV infection.8,10 Given that the hosts for PDX model generation typically exhibit defective innate and adaptive immunity, these human B-cell derived lymphomas can be regarded as animal models of EBV-induced lymphomas in immunocompromised environments.8-12
To understand the pathogenesis of EBV-induced lymphomas in immune deficiency, we explored genomic and transcriptomic alterations through an integrated data analysis from whole-exome sequencing and RNA sequencing (RNA-seq) of human-derived lymphoma in PDX models. Mutations and altered gene expressions in the Rho pathway were found and then strongly suggested as druggable candidate targets in EBV+-DLBLs, using in vivo PDX cases.
Methods
Additional details for methods can be found in supplemental Methods, available on the Blood Web site.
Generation of PDX models
Gastric cancer tissue samples were obtained from individuals who underwent gastrectomies at Seoul National University Hospital in 2014 and 2015. All samples were obtained with informed consent at the Seoul National University Hospital, and the study was approved by the institutional review board of Seoul National University Hospital (No. C-1402-054-555) in accordance with the Declaration of Helsinki. Mice were cared for according to institutional guidelines of the Institutional Animal Care and Use Committee of Seoul National University Hospital (No. 14-0016-C0A0). For PDX models, surgically resected tissues were minced into pieces approximately 2 mm in size and injected into the subcutaneous area in the flanks of 6-week-old nonobese diabetic/severe combined immunodeficiency/interleukin 2γ-receptor null female mice (The Jackson Laboratory). The tumor volume and body weight of the mice were checked once or twice weekly. The volume was calculated as (length × width2)/2. When tumor volumes reached >700 to 1000 mm3, mice were sacrificed and tumor tissues excised and stored. Lymphomas or lymphoproliferative disorders were classified according to the 4th World Health Organization classification.13
High-throughput sequencing and data analysis
Constructed DNA libraries that passed quality checks were loaded onto the HiSeq2000 platform (Illumina Inc.) for paired-end sequencing. Sequenced reads were filtered using the mouse genome (mm9) and then aligned to the human reference genome (hg19), using Burrows-Wheeler Aligner14 with default settings. Demultiplexing was performed using MarkDuplicates in the Picard package to remove polymerase chain reaction (PCR) duplicates. De-duplicated reads were realigned at known indel positions, using GATK IndelRealigner, and the base quality was recalibrated using GATK TableRecalibration.15-17 Somatic variant calling for single-nucleotide variants and short indels was performed with matched normal tissue, using MuTect.18 Final somatic variants were annotated using annotate variation (ANNOVAR).19 RNA-seq was performed using the HiSeq2000 platform (Illumina Inc.), and RNA-seq data analysis was performed using the TCGA RNAseq Version 2 pipeline for mapping with MapSplice20 and gene expression estimation with RSEM.21 Expression signatures were defined as mean values of the expression of related genes (supplemental Table 1). For EBV gene expression, EBV whole-genome sequences were downloaded from NCBI (https://www.ncbi.nlm.nih.gov), and repeated lesions were masked. Next, FASTQ files from RNA-seq were mapped to this reference genome, using Tophat v2.0.7.22 Finally, transcripts per million for each gene was calculated as a measure of gene expression.
Immunohistochemistry and EBV in situ hybridization
Four-micrometer-thick formalin fixed, paraffin-embedded tissue sections were transferred onto silanized charged slides and allowed to dry for 10 minutes at room temperature, followed by 20 minutes in an incubator at 65°C. Sections underwent heat-induced epitope retrieval, using Cell Conditioning 1 buffer for 24 minutes/32 minutes and incubated for 16 minutes with anti-LAMBDA (clone N10/2; 1:2000, Dako), anti-KAPPA (clone R10-21-F3; 1:10 000, Dako), anti-Ki-67 (clone MIB1; 1:200, Dako), anti-CD20 (clone M0755, 1:400, Dako), and anti-CD3 (rabbit polyclonal, 1:100, Dako) in an autoimmunostainer. Antigen-antibody reactions were visualized using a Ventana OptiView DAB IHC Detection Kit (Ventana Medical Systems). Counterstaining was performed using Ventana Hematoxylin II for 12 minutes, and bluing reagent for 4 minutes. EBV was detected in paraffin sections, using a BenchMark XT automatic immunostaining device (Ventana Medical Systems) according to the manufacturer's instructions.
Immunoglobulin H gene clonality assay
Immunoglobulin H (IgH) gene clonality testing was performed using an IdentiClone IGH Gene Clonality Assay Kit (InVivoScribe Technologies). Testing was performed in a pathology laboratory approved by the International Organization for Standardization, using in-house standard protocol (ref. no. AMCP-ISO-QI-2002-M109002[02]).
Rho activity assays
Rho activity was determined in HEK293 cells using specific pull-down assays for the activated forms of the Rho protein, according to the manufacturer’s instructions (Active Rho Detection Kit; Cell Signaling Technology). Briefly, cells were lysed in Lysis/Binding/Wash buffer, and Rhotekin-RBD bound to glutathione-agarose beads was used for affinity precipitation of GTP-bound Rho from the cell lysates. GTP-bound Rho (active Rho), and total Rho were resolved by SDS-PAGE and detected by western blotting, using anti-Rho rabbit antibodies.
Droplet digital PCR
Genomic DNA was restricted with EcoRI (New England Biolabs) for 1 hour at 37°C. The PCR mixture comprised a 20-μL solution containing 1× droplet digital PCR (ddPCR) supermix (Bio-Rad), 1× probe and primer premix for determining wild-type and mutant RHPN2 (at a final concentration of 250 nM for the probe and 900 nM for each primer; Applied Biosystems), and 10 ng of the restricted DNA. The reaction mixture and droplet generation oil (Bio-Rad) were loaded into the droplet generator (QX-200; Bio-Rad). The droplets were transferred to a 96-well PCR plate, and the PCR reaction was performed as follows: 40 cycles of 94°C for 30 seconds, 60°C for 1 minute, and 98°C for 10 minutes. The PCR plate was placed in a droplet reader (Bio-Rad). After the reading, the allele frequencies of mutant RHPN2 were analyzed using Quanta software (Bio-Rad) provided with the droplet reader.
In vivo pharmacological studies
For PDX mice, drug treatments began after the tumors reached approximately 200 mm3. Mice were divided randomly into control and fasudil-treated groups, with 5 mice in each group. Fasudil (Selleckchem, 50 mg/kg, daily) in saline was administered via intraperitoneal injection for 17 days. CHOP (cyclophosphamide, hydroxydaunorubicin, Oncovin, and prednisone) treatment is as follows: cyclophosphamide, 30 mg/kg IV, day 1; hydroxydoxorubicin, 2.475 mg/kg IV, day 1; vincristine, 0.375 mg/kg IV, day 1; and prednisone, 0.15 mg/kg by mouth, days 1-5. Tumor volume was evaluated 3 times weekly and calculated as (length × width2)/2.
Results
Development of EBV+-DLBLs in PDX models of gastric adenocarcinomas
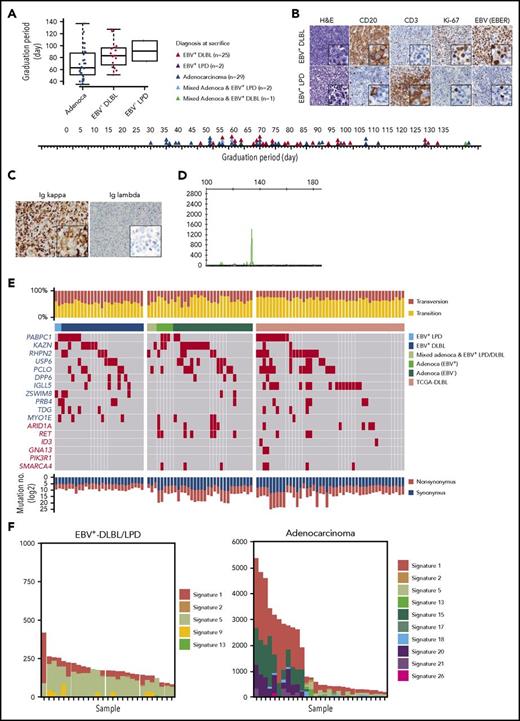
While developing PDX mouse models of gastric cancer, we observed the growth of a significant number of lymphoid cells instead of the intended gastric adenocarcinomas in immunocompromised mice. Of the 59 adenocarcinoma implantations, 25 developed into EBV+-DLBLs, 2 developed into EBV-positive lymphoproliferative diseases (EBV+-LPDs) with marked T-cell expansion, 2 resulted in mixed adenocarcinoma and EBV+-LPD, 1 resulted in a mixed adenocarcinoma and EBV+-DLBL, and 29 resulted in gastric adenocarcinomas with no observable lymphoma/LPD. Compared with adenocarcinoma development, lymphoma development tended to be prolonged (Figure 1A). In histological analysis, all EBV+-DLBLs were positive for CD20 and had a high Ki-67 labeling index (Figure 1B), characteristic of high-grade B-cell lymphoma, and all DLBLs and LPDs from PDXs were EBV-positive, as assessed by EBV-encoded RNA in situ hybridization (EBV-ISH; Figure 1B). The representative 8 samples showed monoclonal EBV type with respect to EBNA2 and LMP1 gene (supplemental Figure 1). In the EBV+-LPD samples, T cells did not show TCR gene rearrangement, suggesting a polyclonal T-cell population (supplemental Figure 2). Clinical characteristics including tumor size, classification, and tumor stage of primary tumors were not associated with EBV+-DLBL development in PDX tumors (supplemental Table 2). RNA-seq analysis for EBV transcript and EBV-ISH in human primary gastric adenocarcinomas showed that tested all primary gastric cancer tissues that developed adenocarcinoma PDX tumors were negative for EBV, and only 2 primary gastric cancer samples that developed DLBL PDX tumors were positive for EBV (supplemental Table 3; supplemental Figure 3). These results suggest that EBV status in primary patient tumors was not associated with EBV+-DLBL development, and that active EBV replication began after tumors were implanted in mice because EBVs were undetected in the majority of primary tumor samples by EBV-ISH. All 25 EBV+-DLBL samples showed κ light-chain restriction by immunohistochemistry (Figure 1C) and were positive for clonal immunoglobulin heavy chain gene rearrangement by IgH gene clonality assay, indicating the presence of a clonal cell population (Figure 1D).
Development of human EBV+-DLBLs from PDXs of gastric cancers. (A) Timeline of the development of EBV+-DLBL, LPD with marked T-cell expansion, and gastric adenocarcinoma in 59 gastric cancer PDX mouse models. Engrafted tumors were considered to have graduated when the tumors reached about 1000 mm3 in size. (B) Immunohistological analysis of EBV+-DLBL and EBV+-LPD that developed in gastric cancer PDXs. Brown color indicates a positive result for CD20, CD3, Ki-67, and EBV-encoded RNA (EBER). All developed lymphomas were CD20-positive, showed high proliferation based on the Ki-67 labeling index (> 95%), and were judged EBV-positive by in situ hybridization. All LPDs showed rich CD3-positive lymphocyte infiltration with a few EBV-positive B cells. (C) Lymphoma cells showed diffuse positivity for immunoglobulin κ chain, but negativity for λ chain in immunohistochemistry. (D) Lymphoma showed clonal proliferation in the IgH gene rearrangement test. (E) Somatic mutation profiles of 59 PDX samples and 48 DLBLs from the TCGA database. Genes marked in red text are frequently altered in Burkitt lymphoma. (F) Mutation signatures observed in EBV+-DLBL and gastric adenocarcinoma PDX samples. The x-axes represent each sample and were ordered according to the total number of somatic substitutions (y-axes). Contributions of each mutational signature are shown as bar graphs.
Development of human EBV+-DLBLs from PDXs of gastric cancers. (A) Timeline of the development of EBV+-DLBL, LPD with marked T-cell expansion, and gastric adenocarcinoma in 59 gastric cancer PDX mouse models. Engrafted tumors were considered to have graduated when the tumors reached about 1000 mm3 in size. (B) Immunohistological analysis of EBV+-DLBL and EBV+-LPD that developed in gastric cancer PDXs. Brown color indicates a positive result for CD20, CD3, Ki-67, and EBV-encoded RNA (EBER). All developed lymphomas were CD20-positive, showed high proliferation based on the Ki-67 labeling index (> 95%), and were judged EBV-positive by in situ hybridization. All LPDs showed rich CD3-positive lymphocyte infiltration with a few EBV-positive B cells. (C) Lymphoma cells showed diffuse positivity for immunoglobulin κ chain, but negativity for λ chain in immunohistochemistry. (D) Lymphoma showed clonal proliferation in the IgH gene rearrangement test. (E) Somatic mutation profiles of 59 PDX samples and 48 DLBLs from the TCGA database. Genes marked in red text are frequently altered in Burkitt lymphoma. (F) Mutation signatures observed in EBV+-DLBL and gastric adenocarcinoma PDX samples. The x-axes represent each sample and were ordered according to the total number of somatic substitutions (y-axes). Contributions of each mutational signature are shown as bar graphs.
Mutational analysis of EBV+-DLBLs from PDX models
Somatic mutation analysis was performed in all 59 tumors derived from gastric adenocarcinoma implantation (supplemental Table 4); the results were also compared with sequencing data of DLBLs from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov) and of 59 Burkitt lymphoma cases23 (Figure 1E; supplemental Figure 4). In all DLBLs from TCGA (TCGA-DLBLs), except for 1 case, EBV viral RNA was not detected from RNA-seq data. The total number of mutations per sample in the 25 EBV+-DLBLs and 2 EBV+-LPDs (mean, 143.3; standard deviation [SD], 11.9) was significantly lower than that in the gastric adenocarcinomas (mean, 1482.2; SD, 294.4; P = .0001) and the TCGA-DLBLs (mean, 1477.9; SD, 418.5; P = .0026). Transversion mutations occurred at a higher rate in EBV+-DLBLs (mean, 0.45; SD, 0.06) than in adenocarcinomas (mean, 0.34; SD, 0.12; P = .0003) or TCGA-DLBLs (mean, 0.30; SD, 0.06; P < .0001) (Figure 1E). In addition, the overall mutational patterns of EBV+-DLBLs were distinct from those of adenocarcinomas (supplemental Figure 5). Furthermore, the mutational profiles of EBV+-DLBLs were markedly different from matched primary gastric adenocarcinoma samples from patients (supplemental Figure 6), precluding the possibility of selecting preexisting mutated gastric cancer cells and suggesting the de novo emergence of these mutations. For gastric adenocarcinoma PDX models, EBV positivity was detected in 5 PDX cases (PDX adenocarcinoma with EBV+) by EBV-ISH, but EBV was only present in background lymphocytes around cancer cells (supplemental Figure 7). Little difference in mutation burden and mutation profiles was observed between EBV+- and EBV−-adenocarcinomas of PDX models (supplemental Figure 8).
Investigating the mutation signatures, based on the trinucleotide context of sequence mutations24 (a curated list of mutational signatures and their characteristics can be on the Catalogue of Somatic Mutations in Cancer [COSMIC] signature website: http://cancer.sanger.ac.uk/cosmic/signatures), showed that the signatures in the different EBV+-DLBL/LPD samples (n = 27) were similar to one another but were remarkably different from those of adenocarcinoma samples (Figure 1F). In the EBV+-DLBL/LPD samples, signatures 5 (unknown etiology) and 1 (spontaneous 5-methylcytosine deamination) were dominant, with minor contributions from signatures 2, 13 (both 2 and 13 are attributed to APOBEC-related mutagenesis), and 9 (polymerase η-related activation-induced cytidine deaminase activity). Signature 5 is a broad spectrum of base changes with transcriptional strand bias. Signatures 5 and 1 are frequently seen in the majority of cancers and normal tissues,25 and the number of mutations in a cell attributable to these processes is proportional to the chronological age of the individual, suggesting that the underlying mutational processes are endogenous and active throughout life at a constant rate (ie, clocklike property).24 These 5 mutational signatures are all found in the genomes of primary B-cell lymphomas,24 suggesting that no other artificial mutational processes had been activated during the formation of PDX mouse models.
The frequently altered somatic mutations in our EBV+-DLBL/LPD samples were in the PABPC1, KAZN, RHPN2, USP6, PCLO, and DPP6 genes; this differed from the mutational features of Burkitt lymphoma (frequent mutations in ARIDA, RET, ID3, GNA13, PIK3R1, and SMARCA4) and TCGA-DLBLs (Figure 1E; supplemental Figure 4). Interestingly, the KAZN, RHPN2, and USP6 genes, which were frequently mutated in EBV+-DLBL (Figure 1E; 60%, 15/25 cases), are involved in the Rho pathway.26-28 One EBV+-DLBL sample from TCGA also showed mutations in Rho pathway-associated genes, including KTN1, PKN2, CDH3, MAP1B, and ARHGAP23 (supplemental Table 5). The Rho pathway mainly regulates cytoskeletal and cell adhesion dynamics, and thereby participates in a variety of cellular processes, including cell polarity, migration, and cell cycle progression. Mutations in KAZN, RHPN2, and USP6 genes were validated using Sanger sequencing (supplemental Figure 9; supplemental Table 6). However, mutations in KAZN, RHPN2, and USP2 found in EBV+-DLBL PDX samples were not detected in Sanger sequencing of matched primary gastric adenocarcinoma tissues from patients (supplemental Figure 10).
Alterations of Rho pathway by RHPN2 mutations in EBV+-DLBLs
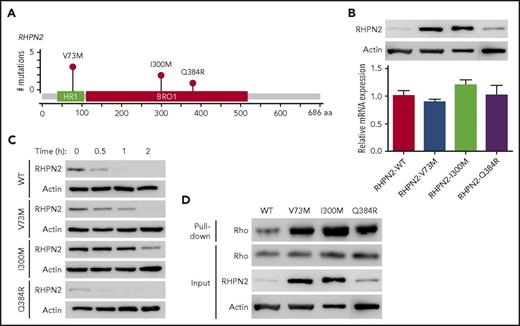
Rhophilin 2 (RHPN2), identified as a binding partner of RhoA GTPase,27 is associated with mesenchymal transformation, as it activates RhoA and induces cell invasion in malignant glioma.29 In our EBV+-DLBL samples, RHPN2 mutations were detected in 17.2% (5/29) of the cases sequenced, and we found 6 RHPN2 mutations in 5 patients (Figure 2A). Among them, the V73M and I300M mutations were found recurrently in 3 and 2 patients, respectively (Figure 2A). Recurrent V73M mutations in RHPN2 were previously reported in Asian patients with lung adenocarcinoma,30 and I300M mutations have been reported in the COSMIC database (http://cancer.sanger.ac.uk/cosmic).
Activation of Rho signaling by mutant RHPN2 proteins. (A) Mutation needle plots of RHPN2 in EBV+-DLBL. The mutations found in the 5 EBV+-DLBLs of 29 cases sequenced are shown. BRO1, yeast Bro1 homology domain; HR1, protein kinase C-related kinase homology region 1 domain. (B) Protein levels of wild-type and mutant RHPN2 when mRNA levels were normalized. (C) Protein stability of wild-type and mutant RHPN2. The protein levels of each mutant RHPN2 were evaluated after 3 hours of treatment with cycloheximide (100 μg/mL). (D) Rho activity of wild-type and mutant RHPN2. Rho activity were assessed by a GST-RBD-Rhotekin pull-down assay, and the RHPN2 mutations were associated with increased Rho activity.
Activation of Rho signaling by mutant RHPN2 proteins. (A) Mutation needle plots of RHPN2 in EBV+-DLBL. The mutations found in the 5 EBV+-DLBLs of 29 cases sequenced are shown. BRO1, yeast Bro1 homology domain; HR1, protein kinase C-related kinase homology region 1 domain. (B) Protein levels of wild-type and mutant RHPN2 when mRNA levels were normalized. (C) Protein stability of wild-type and mutant RHPN2. The protein levels of each mutant RHPN2 were evaluated after 3 hours of treatment with cycloheximide (100 μg/mL). (D) Rho activity of wild-type and mutant RHPN2. Rho activity were assessed by a GST-RBD-Rhotekin pull-down assay, and the RHPN2 mutations were associated with increased Rho activity.
We then investigated the functional changes resulting from the RHPN2 mutations, including their effects on Rho pathway activity. The RHPN2 V73M and I300M mutations appeared to increase protein levels despite having little effect on mRNA levels (Figure 2B). In addition, the V73M and I300M mutant proteins showed prolonged half-lives (Figure 2C), possibly resulting from increased protein stabilization of the mutant protein. The increased levels of RHPN2 mutant proteins were accompanied by increased Rho activity (Figure 2D). The mutant RHPN2 protein I300M was also predicted to have changes in its protein secondary structure by in silico protein structure prediction (supplemental Figure 11), suggesting that the change in secondary structure may have implications for the protein’s prolonged half-life.
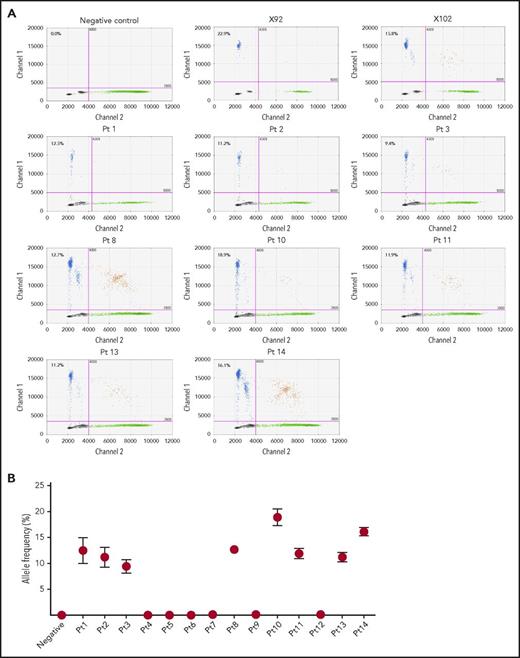
We identified RHPN2 I300M mutations in human EBV+-DLBL tumor tissues from 8 of 14 additional unrelated patients (57.1%), using ddPCR and Sanger sequencing (Figure 3; supplemental Figure 12A), and the presence of EBV in these human patient tumors was confirmed by immunohistological analysis (supplemental Figure 13). Among the 14 patient samples, 11 cases were EBV+-DLBL of the elderly and 3 cases were associated with transplantation (posttransplant lymphoproliferative disease; detailed clinical information in supplemental Table 7), but the rate of RHPN2 mutations was not significantly different between the 2 groups (P = .209; supplemental Table 7). We also detected RHPN2 I300M mutations in 24 of 52 EBV-transformed lymphoblastoid cell lines (LCLs; 46.2%), using ddPCR (supplemental Figure 14). The representative LCL samples showed IgH gene rearrangement regardless of RHPN2 mutation status (supplemental Figure 15)., and the expression of human telomerase reverse transcriptase was not significantly different between LCLs with wild-type RHPN2 and mutant RHPN2 (P = .5349; supplemental Figure 16), suggesting that the telomere stabilization in LCLs was not associated with the RHPN2 mutation status. Furthermore, 1 patient had an additional RHPN2 mutation (Q384R) and a mutation in USP6 (R133K), another gene in the Rho pathway, which were also detected in our EBV+-DLBL samples generated in PDXs (supplemental Figure 12B-C), suggesting that the mutational profiles of EBV+-DLBL samples generated in our PDXs do indeed reflect the mutations observed in human EBV+-DLBL patients.
Detection of RHPN2 mutations in human EBV+-DLBL samples. (A) Representative graphs of the detection of RHPN2 mutation (I300M), using ddPCR. Multiplex ddPCR was performed for wild-type RHPN2 (green dots) and mutant RHPN2 (I300M; blue dots), using blood (negative control) and tumor DNA from human EBV+-DLBL samples. The numbers in the graphs indicate the allele frequency of mutant RHPN2. X92 and X102 were EBV+-DLBL from PDX models, which were positive controls for the I300M mutation. Pt, patient. (B) Estimation of allele frequencies of RHPN2 I300M mutations in 14 human EBV+-DLBL samples using ddPCR. Error bars indicate the Poisson distribution at the 95% confidence interval.
Detection of RHPN2 mutations in human EBV+-DLBL samples. (A) Representative graphs of the detection of RHPN2 mutation (I300M), using ddPCR. Multiplex ddPCR was performed for wild-type RHPN2 (green dots) and mutant RHPN2 (I300M; blue dots), using blood (negative control) and tumor DNA from human EBV+-DLBL samples. The numbers in the graphs indicate the allele frequency of mutant RHPN2. X92 and X102 were EBV+-DLBL from PDX models, which were positive controls for the I300M mutation. Pt, patient. (B) Estimation of allele frequencies of RHPN2 I300M mutations in 14 human EBV+-DLBL samples using ddPCR. Error bars indicate the Poisson distribution at the 95% confidence interval.
Transcriptomic activation of Rho pathway in EBV+-DLBLs
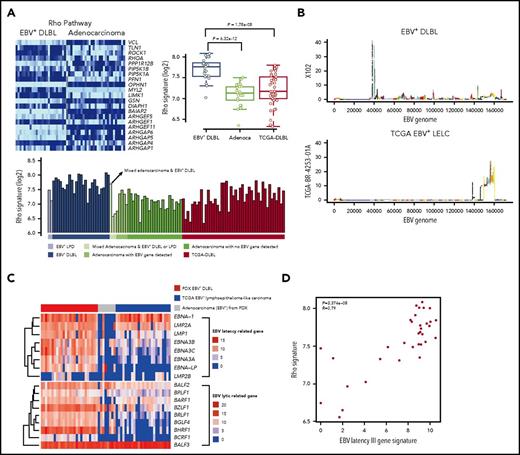
To investigate gene expression alterations in EBV+-DLBLs, we performed transcriptomic analysis of tumor tissues, using RNA-seq data (supplemental Table 8). The expression pattern of EBV+-DLBLs in this study differed from those of adenocarcinomas and EBV-negative TCGA-DLBLs, as assessed by nonnegative matrix factorization and principal component analysis (supplemental Figure 17A). When the immune signatures of the tumor tissues were investigated using same RNA-seq data, all EBV+-DLBL tissues showed an increased plasma cell signature, whereas half of the EBV-negative TCGA-DLBLs and all gastric adenocarcinoma (no lymphoma development) cases showed a decreased plasma cell signature (supplemental Figure 17B). When the immunostaining for IgA and IgG was tested in tumor tissues, we identified that the main source of plasm call signature was infiltrating plasma cells in tumor background (supplemental Figure 17C), suggesting that infiltrating plasma cells may have a role in lymphoma development or a trace of humoral immune reaction for EBV, because there was little plasma cell infiltration in adenocarcinoma. The plasma cell signature correlated with IRF4 expression level (supplemental Figure 18), which is important in the regulation of interferons in response to viral infection. Pathway analysis using the transcriptome data showed that the Rho pathway was significantly altered in EBV+-DLBLs compared with adenocarcinomas and EBV-negative TCGA-DLBL samples (Figure 4A; supplemental Table 9).
Transcriptomic alterations in EBV+-DLBL from PDX models. (A) Increased Rho pathway signature in EBV+-DLBL from PDX models. Alterations of the Rho pathway were identified by pathway analysis using 3 tumor types (EBV+-DLBLs, gastric adenocarcinomas, and DLBLs from the TCGA database). (B) EBV gene expression activity across the entire EBV genome. The EBV viral gene activation pattern differed between EBV+-DLBLs from our study (X102 is representative) and EBV-associated gastric adenocarcinomas from the TCGA database (TCGA BR-4253-01A is representative). (C) Expression profiles of latency and lytic cycle-related EBV genes. EBV+-DLBL showed a latency III pattern in gene expression, whereas limited latency-related gene expression was present in EBV+ gastric adenocarcinoma. Lytic cycle-related genes were expressed in all 3 lytic cycle stages in EBV+-DLBLs, but gene expression was limited in TCGA adenocarcinoma. (D) Correlation between the EBV latency III-related gene signature and Rho pathway signature. Each dot represents an EBV+-DLBL sample, and the numbers in the graph indicate P-value and Pearson correlation coefficient (R) of the correlation.
Transcriptomic alterations in EBV+-DLBL from PDX models. (A) Increased Rho pathway signature in EBV+-DLBL from PDX models. Alterations of the Rho pathway were identified by pathway analysis using 3 tumor types (EBV+-DLBLs, gastric adenocarcinomas, and DLBLs from the TCGA database). (B) EBV gene expression activity across the entire EBV genome. The EBV viral gene activation pattern differed between EBV+-DLBLs from our study (X102 is representative) and EBV-associated gastric adenocarcinomas from the TCGA database (TCGA BR-4253-01A is representative). (C) Expression profiles of latency and lytic cycle-related EBV genes. EBV+-DLBL showed a latency III pattern in gene expression, whereas limited latency-related gene expression was present in EBV+ gastric adenocarcinoma. Lytic cycle-related genes were expressed in all 3 lytic cycle stages in EBV+-DLBLs, but gene expression was limited in TCGA adenocarcinoma. (D) Correlation between the EBV latency III-related gene signature and Rho pathway signature. Each dot represents an EBV+-DLBL sample, and the numbers in the graph indicate P-value and Pearson correlation coefficient (R) of the correlation.
We then explored the EBV viral gene expression in EBV+-DLBLs to investigate the latency status of EBV (supplemental Table 10). Using the same RNA-seq data as earlier, we detected EBV (human herpesvirus 4) viral mRNA in all EBV+-DLBLs from our PDXs, with the exception of 1 case, which was positive for EBV via in situ hybridization (supplemental Figure 19A). In contrast, we showed that, with the exception of 1 case, all DLBLs from the TCGA database were negative for EBV, based on RNA-seq data (supplemental Figure 19B). Interestingly, EBV viral mRNA was also detected in several gastric adenocarcinoma PDX samples that did not develop into lymphomas (PDX adenocarcinoma with EBV+; supplemental Figure 19A), suggesting that the presence of viral mRNA alone is insufficient to induce lymphomagenesis. To compare the EBV viral gene-expression patterns between lymphoma and epithelial carcinoma, we downloaded gene expression data from 25 EBV-associated lymphoepithelioma-like carcinomas (LELCs) of stomach from the TCGA database (TCGA-gastric EBV+-LELC), because EBV is positive in epithelial carcinoma cells of this tumor type. EBV viral gene expression patterns were distinctively different between EBV+-DLBLs from the present study and TCGA-gastric EBV+-LELC (Figure 4B; supplemental Figure 20). Primary EBV infection typically shows 3 distinct latency types (latency I, II, and III), depending on the viral gene expression pattern.31 Our EBV+-DLBLs exhibited a type III latency state, in which most EBV latent genes are expressed (Figure 4C). This result correlates well with the latency pattern in posttransplantation lymphoproliferative disease and in vitro EBV infection-mediated establishment of lymphoblastoid cell lines.31 However, limited and heterogeneous EBV latent gene expression was present in our PDX adenocarcinomas with EBV+ (Figure 4C). These findings suggest that EBV viral gene expression differs between EBV-infected lymphocytes and EBV-infected epithelial cells, and that EBV infection in EBV+-DLBLs is characterized by the predominance of an EBV latency III signature. When the lytic cycle-related gene expression pattern was also evaluated, the expression pattern was again different between EBV+-DLBLs and TCGA-gastric EBV+-LELCs (Figure 4C). Lytic genes in all 3 stages (immediate-early, early, and late) were expressed in most EBV+-DLBLs. This finding correlated well with the result that most EBV+-DLBL showed plasma cell signature because plasma cell differentiation is supposed to be associated with the EBV lytic cycle. A limited lytic gene expression pattern was present in TCGA-gastric EBV+-LELC (Figure 4C).
Next, we investigated the association between the EBV latency III gene signature and human transcriptomic changes in EBV+-DLBL cells to investigate the pathogenesis of EBV-induced DLBLs (supplemental Figure 21). The top 5 human genes that most closely correlated with the EBV latency III signature were CLEC17A, MS4A1 (CD20), NFKBID, PRPSAP2, and CD22 (correlation coefficient >0.9); of note, NFKBID is p100, which can evoke the alternative NF-κ B pathway. Pathway analysis using correlated genes of correlation coefficient >0.75 highlighted that the NF-kB signaling pathway and the Toll-like receptor signaling pathway were altered (supplemental Figure 21), suggesting that activation of NF-kB signaling pathway and the Toll-like receptor signaling pathway is associated with EBV-induced lymphomagenesis by EBV latency III genes. In addition, we also observed that the EBV latency III signature was significantly correlated with the Rho pathway gene signature (P = 3.374 × 10−8; Figure 4D).
Efficacy of Rho pathway-targeting agent in in vivo PDX models of EBV+-DLBL
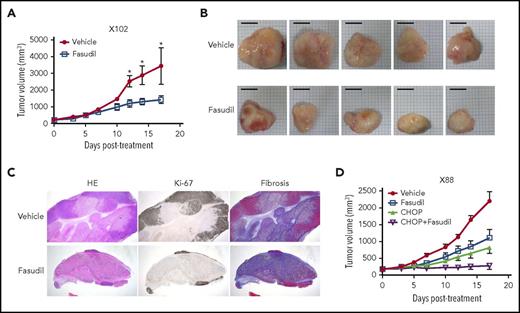
We then investigated the feasibility of targeting the Rho pathway for the treatment of EBV-induced lymphomas, because we identified Rho pathway alterations in both genomic and transcriptomic analyses (Figure 1E; Figure 4A). Rho-associated protein kinase (ROCK) is a major downstream effector of Rho, and fasudil is a clinically approved ROCK inhibitor for the treatment of vasospasm.32 When fasudil was applied to 2 different PDX models of EBV-induced lymphomas (1 with the RHPN2 I300M mutation [X102] and the other with transcriptional activation of the Rho pathway [but no RHPN2 mutation; X158]), the fasudil-treated group showed significantly decreased tumor growth compared with the vehicle-treated samples (Figure 5A; supplemental Figure 22A). Morphological evaluation showed decreased tumor sizes in the fasudil treatment groups (Figure 5B; supplemental Figure 22B). Histologically, tumors treated with fasudil showed decreased numbers of viable tumor cells, with increased tumor necrosis replaced by fibrosis (Figure 5C). In an in vitro experiment, LCLs with RHPN2 I300M mutations (AL073 and AL095) were more sensitive to fasudil than LCLs with wild-type RHPN2 (AL291 and AL817; supplemental Figure 23), suggesting that fasudil was more effective in cells with RHPN2 I300M mutations. The growth of engrafted tumors of LCL with RHPN2 I300M mutations (AL073) was also inhibited by fasudil treatment (supplemental Figure 24). In addition, the combination treatment of fasudil and a standard regimen, CHOP, showed a synergistic effect in another PDX case, with the RHPN2 V73M mutation (X88; Figure 5D; supplemental Figure 22C).
In vivo efficacy of a ROCK inhibitor, fasudil, in EBV+-DLBL mouse models. (A-B) The efficacy of a ROCK inhibitor, fasudil, in EBV+-DLBL mouse models with the RHPN2 I300M mutation (X102). Mice were treated with fasudil (50 mg/kg/day) or vehicle for 17 days (n = 5). Average tumor sizes for each group were plotted (A), and representative tumors after treatment are shown (B). Asterisks indicate statistically significant differences (*P < .05) between vehicle- and fasudil-treated groups. Scale bar, 10 mm. (C) Immunohistological analysis of fasudil-treated residual tumors in EBV+-DLBL PDX models. Microscopic examination (hematoxylin and eosin stain, 1.25×) showed decreased numbers of viable cells highlighted by Ki-67 labeling and increased necrosis with fibrosis. Fibrosis was determined by collagen staining. HE, hematoxylin and eosin stain. (D) The synergistic effect of fasudil and CHOP in EBV+-DLBL mouse models with the RHPN2 V73M mutation (X88). Mice were treated with fasudil (50 mg/kg/day), CHOP (cyclophosphamide, 30 mg/kg, day 1; hydroxydoxorubicin, 2.475 mg/kg, day 1; vincristine, 0.375 mg/kg, day 1; and prednisone, 0.15 mg/kg, day 1-5), or a combination of fasudil and CHOP for 17 days (n = 5), and average tumor sizes for each group were plotted.
In vivo efficacy of a ROCK inhibitor, fasudil, in EBV+-DLBL mouse models. (A-B) The efficacy of a ROCK inhibitor, fasudil, in EBV+-DLBL mouse models with the RHPN2 I300M mutation (X102). Mice were treated with fasudil (50 mg/kg/day) or vehicle for 17 days (n = 5). Average tumor sizes for each group were plotted (A), and representative tumors after treatment are shown (B). Asterisks indicate statistically significant differences (*P < .05) between vehicle- and fasudil-treated groups. Scale bar, 10 mm. (C) Immunohistological analysis of fasudil-treated residual tumors in EBV+-DLBL PDX models. Microscopic examination (hematoxylin and eosin stain, 1.25×) showed decreased numbers of viable cells highlighted by Ki-67 labeling and increased necrosis with fibrosis. Fibrosis was determined by collagen staining. HE, hematoxylin and eosin stain. (D) The synergistic effect of fasudil and CHOP in EBV+-DLBL mouse models with the RHPN2 V73M mutation (X88). Mice were treated with fasudil (50 mg/kg/day), CHOP (cyclophosphamide, 30 mg/kg, day 1; hydroxydoxorubicin, 2.475 mg/kg, day 1; vincristine, 0.375 mg/kg, day 1; and prednisone, 0.15 mg/kg, day 1-5), or a combination of fasudil and CHOP for 17 days (n = 5), and average tumor sizes for each group were plotted.
Discussion
Lymphomagenesis of human tumors in immune-deficient mice has been reported in PDX models of several cancer types, including non-small cell lung cancers, hepatocellular carcinomas, prostate cancers, and gastric cancers.8-10,33 Most of these lymphomas were CD45-positive B-cell subtypes and were frequently associated with EBV infection.8,10 Inflammation of the original tumor tissue increased this lymphomagenesis, and gastric cancers exhibited a notably higher lymphomagenesis rate, in part because of higher baseline inflammation.10 It was also reported that EBV-positive tumors arising after the engraftment of lymph nodes from patients with Hodgkin lymphoma were derived from bystander-quiescent EBV-positive B lymphocytes and not from the EBV-positive Reed-Sternberg cells.34
In immunocompromised patients, such as those with HIV infections and organ transplantations, EBV plays a carcinogenic role in the development of NHL, including DLBL, Burkitt lymphoma, extranodal NK/T-cell lymphoma of the nasal type, and nasopharyngeal cancer.5 Indeed, the most frequent histological type of posttransplant lymphoproliferative disease is EBV+-DLBL. In the present study, we propose that the EBV+-DLBLs that developed in PDX mice were compatible with human lymphomas that arise in immunocompromised patients for the following reasons: (1) in histological analysis, PDX samples were diagnosed as DLBLs by 4 independent pathologists, and immunohistochemistry for markers, presence of clonal proliferation, and κ chain restriction were compatible with EBV+-DLBLs; (2) EBV+-DLBL was reported to occur in more severely immunocompromised environments compared with Burkitt lymphoma,5 and nonobese diabetic/severe combined immunodeficiency/interleukin 2γ-receptor null mice represent such severe immunocompromised conditions; (3) in mutational analysis, the mutation signatures of our models, based on the trinucleotide context of sequence mutations, were compatible with the patterns reported in B-cell lymphomas24 ; (4) RHPN2 mutations in our models were detected in 57.1% of unrelated EBV+-DLBL patient samples and 46.2% of EBV-transformed LCLs (Figure 3; supplemental Figure 14); (5) 1 EBV+-DLBL sample from TCGA showed mutations in Rho pathway-associated genes including KTN1, PKN2, CDH3, MAP1B, and ARHGAP23; and (6) in RNA-seq analysis, EBV genes associated with latency III status were expressed in our samples, and expression of EBV latency III gene is characteristic of EBV virus activation and highly associated with EBV-induced lymphomagenesis. Taken together, these findings suggest that EBV+-DLBL in PDX mice represents a valuable experimental model for EBV-positive lymphoma research.
The RHPN2 gene is located at 19q13.11. However, there are also 2 near-identical and truncated sequences on chromosomes 15 and 16, suggesting that the segment is often duplicated in human populations. However, mRNA transcription from the duplicated gene sequences lack exons 1-3, and therefore can be differentiated from the original RHPN2 gene. Our exome sequencing of tumor and matched normal tissues clearly shows that the RHPN2 mutations are tumor specific (therefore somatic, with a ∼50% variant allele fraction), and were detected not in the duplicons but in the bona fide RHPN2 sequences. The mutation is also transcribed and detected from RNA-seq, corroborating our conclusions. However, in the ddPCR validation experiments, which is less specific than exome or RNA sequencing, a low variant allele frequency of the mutations are strongly expected because PCR amplicons will also be generated from the 2 duplicated segments (expected to be ∼17%, 1 mutation copy of a total of 6 copies). Therefore, a seemingly low variant allele frequency in ddPCR experiments does not mean subclonality of the mutation in the cancer cells. Indeed, because our validation results are close to the theoretical expectation (∼17%), we actually believe that the mutations are all clonal in the EBV+-DLBL samples from patient cancer tissues and PDX tumors. And a low variant allele frequency in patient samples seems to be also associated with tumor cellularity in lymphoma tissues, because there was an infiltration of nonneoplastic lymphoplasma cells.
Our exome and transcriptome analyses suggest that the genomic and transcriptomic alterations of the Rho pathway might serve as novel oncogenic drivers in EBV-induced lymphomagenesis in an immune compromised status. A type III latency reprogramming, in which most EBV latent genes are expressed, is essential for EBV-induced B-cell transformation and is highly associated with the Rho pathway gene expression signature (Figure 4D). In a subset of cases, occurrence of mutations in genes of the Rho pathway such as RHPN2, KAZN, and USP6 plays roles cooperatively with lymphomagenesis. The importance of genomic and transcriptomic activation of Rho pathway in lymphomagenesis was verified in PDX experiments, in which inhibition of Rho pathway with a ROCK inhibitor, fasudil, reduced tumor burden not only in samples with RHPN2 mutations (X102, X88) but also in sample with transcriptional activation of the Rho pathway (but no RHPN2 mutation; X158). These alterations can be found in the context of EBV activation, which is usually linked to immune compromised status. Thus, the detection of RHPN2 mutations and measuring the Rho pathway transcriptional signature could be suggested as a new companion diagnostics for fasudil treatment in EBV+ DLBLs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Stephen Sampson for editing the manuscript and Asis Das for critical reading of the manuscript. This work was supported by the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (grant HI13C2148 and HI16C2387); the International Research & Development Program of the National Research Foundation of Korea, funded by the Ministry of Science, ICT and Future Planning (grant no. 2015K1A4A3047851); the National Research Foundation of Korea grant, funded by the Korea government (grant 2017R1C1B2002183); and the National Cancer Institute of the National Institutes of Health (grant P30CA034196). C.L. is a distinguished Ewha Womans University Professor supported in part by the Ewha Womans University Research grant of 2015-2017.
Authorship
Contribution: S.-Y.C., C.O.S., J.-I.K., and C.L. conceived and designed the experiments; H.-K.P., Y.H.K., D.L., Y.-S.S., S.-H.K., H.-J.L., and H.-K.Y. collected samples and performed clinical data analyses; C.O.S., J.C., D.N., H.K., J.H.C., S.P., Y.S.J., and J.-I.K. performed high-throughput sequencing and data analysis; S.-Y.C., J.L., D.N., W.K., J. Kang, S.M., A.L., E.K., J. Kim, and B.C. performed molecular and animal experiments; C.O.S., C.-S.P., S.P., J.R., M.-S.C, and W.-H.K. performed histological analysis; S.-Y.C., C.O.S., J.C., J. Keck, J.B., E.T.L., W.-H.K., H.P., H.-K.Y., J.-I.K., and C.L. discussed the results; S.-Y.C., C.O.S., J.-I.K., and C.L. wrote the manuscript; and all authors edited and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sung-Yup Cho, Ewha Institute of Convergence Medicine, Ewha Womans University Mokdong Hospital, 1071 Anyangcheon-ro, Yangcheon-gu, Seoul 07985, Korea; e-mail: csybio@ewha.ac.kr; Jong-Il Kim, Department of Biomedical Sciences, Seoul National University College of Medicine, 103 Daehak-ro, Jongno-gu, Seoul 03080, Korea; e-mail: jongil@snu.ac.kr; Charles Lee, The Jackson Laboratory for Genomic Medicine, 10 Discovery Drive, Farmington, CT 06032; e-mail: charles.lee@jax.org.
References
Author notes
S.-Y.C. and C.O.S. contributed equally as first authors.
H.-K.Y., J.-I.K., and C.L. share senior authorship to this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal