In this issue of Blood, Gallipoli et al report that by performing an elegant clustered regularly interspaced short palindromic repeats (CRISPR) screen of FLT3 internal tandem duplication–positive (FLT3-ITD+) acute myeloid leukemia (AML) cells, they have identified that FLT3 inhibition exposes a therapeutically relevant metabolic dependency on glutaminolysis.1
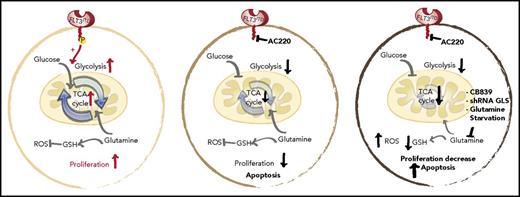
Schematic representation of the glycolytic and glutaminolytic pathways in an FLT3-ITD+ AML cell under normal conditions (left), with FLT3 inhibition (middle), and with FLT3 and glutamine inhibition (right). FLT3 inhibition reduces glycolysis without equally affecting glutamine metabolism or glutathione (GSH) production; thus, combination FLT3/glutamine inhibition synergizes to increases apoptosis in FLT3-ITD cells. P, phospho; ROS, reactive oxygen species; shRNA, short hairpin RNA. See the complete Figure 6 in the article by Gallipoli et al that begins on page 1639.
Schematic representation of the glycolytic and glutaminolytic pathways in an FLT3-ITD+ AML cell under normal conditions (left), with FLT3 inhibition (middle), and with FLT3 and glutamine inhibition (right). FLT3 inhibition reduces glycolysis without equally affecting glutamine metabolism or glutathione (GSH) production; thus, combination FLT3/glutamine inhibition synergizes to increases apoptosis in FLT3-ITD cells. P, phospho; ROS, reactive oxygen species; shRNA, short hairpin RNA. See the complete Figure 6 in the article by Gallipoli et al that begins on page 1639.
One of the hallmarks of cancer is metabolic reprogramming,2 a change that is thought to aid the ability of the cancer to outgrow and outcompete its native untransformed brethren. Extensive metabolic studies of cancer model systems, such as that performed by Gallipoli et al, highlight how this perturbed metabolism can be used against the cancer cells through combination therapy with specific kinase and metabolism inhibitors.
AC220, commonly known as quizartinib, is a second-generation FLT3 inhibitor that exhibits superior potency and selectivity.3 Gallipoli et al use quizartinib as a selective agent in a CRISPR/Cas9 lethality screen of the human AML cell line, MOLM13, to identify novel genes/pathways that facilitate resistance to FLT3 inhibitors. They identify metabolic pathways as the top enriched KEGG gene set enrichment category, with 1 gene known as glutaminase (GLS) exhibiting a particularly strong synthetic lethality.
GLS is the first-stage enzyme in glutamine catabolism, known for its anaplerotic and biosynthesis roles in cancer.4 Previous studies have identified that impairing glutamine metabolism or uptake can exert antileukemic activity.5-7 Gallipoli et al further this line of investigation by providing an in-depth examination of the tricarboxylic acid (TCA) cycle and redox metabolic intermediates (including reduced/oxidized glutathione ratios) in quizartinib-treated FLT3-ITD+ AML cells. Overall, they found that FLT3 inhibition leads to a preferential impairment of glucose utilization and glycolytic pathways, without equally affecting glutamine utilization or metabolism. The unmasked metabolic dependence on glutaminolysis makes pharmacological inhibition of glutamine metabolism a logical combination therapy for FLT3 inhibitor–treated FLT3-ITD+ AML (see figure).
This study provides a strong rationale as to why glutaminase inhibitors in particular lead to significant synergism with FLT3 inhibitors, a concept that is in line with other current studies.8 In addition, the authors ascertain that other oncogenic lesions, such as BCR-ABL, exhibit similar characteristic changes in metabolism. In particular, glycolytic pathways and glycolytic enzyme gene expression were severely hampered in BCR-ABL+ cells after imatinib treatment, whereas other TCA cycle and glutamine metabolism pathways/genes (including GLS) were less affected. This indicates that glutaminolysis, along with other TCA metabolic pathways, may indeed play an important role in increasing the survival and resistance of many oncogenic kinase-driven leukemic cells to targeted kinase inhibition.
Although glutaminolysis contributes a significant proportion of the energy requirements for FLT3-ITD+ cells under FLT3 inhibitory stress, it is not the only metabolic pathway that is functioning. The CRISPR screen by Gallipoli et al reveals that several other genes associated with metabolic pathways demonstrate significant synergistic lethality with quizartinib. These include genes involved in fatty acid synthesis (MECR) and phosphatidylinositol metabolism (PLCH1). Thus, further investigation, similar to that performed here, is warranted in future studies to uncover the role of these other metabolic pathways in providing energy and promoting survival of FLT3-ITD+ AML cells during FLT3 inhibition.
Current treatment of FLT3-ITD+ AML is associated with unacceptably low rates of long-term survival posttherapy.9 The development of specific and potent FLT3 inhibitors offers hope in providing an effective treatment for this aggressive form of AML, but resistance remains a problem. Gallipoli et al provide compelling evidence for a role of glutaminolysis in the development of this resistance, and their data compel further investigation of combination FLT3/glutaminase inhibitor treatment in FLT3-ITD+ patients.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal