Key Points
The intronic A4GALT SNP rs5751348 defines a hematopoietic transcription factor–binding site present in P1 but not P2 blood group alleles.
RUNX1 selectively binds to this regulatory site in P1 alleles; small interfering RNA knockdown of RUNX1 downregulates A4GALT transcript levels.
Abstract
P1 and Pk are glycosphingolipid antigens synthesized by the A4GALT-encoded α1,4-galactosyltransferase, using paragloboside and lactosylceramide as acceptor substrates, respectively. In addition to the compatibility aspects of these histo-blood group molecules, both constitute receptors for multiple microbes and toxins. Presence or absence of P1 antigen on erythrocytes determines the common P1 (P1+Pk+) and P2 (P1−Pk+weak) phenotypes. A4GALT transcript levels are higher in P1 individuals and single-nucleotide polymorphisms (SNPs) in noncoding regions of A4GALT, particularly rs5751348, correlate with P1/P2 status. Despite these recent findings, the molecular mechanism underlying these phenotypes remains elusive. The In(Lu) phenotype is caused by Krüppel-like factor 1 (KLF1) haploinsufficiency and shows decreased P1 levels on erythrocytes. We therefore hypothesized KLF1 regulates A4GALT expression. Intriguingly, P1-specific sequences including rs5751348 revealed potential binding sites for several hematopoietic transcription factors, including KLF1. However, KLF1 binding did not explain P1-specific shifts in electrophoretic mobility-shift assays and small interfering RNA silencing of KLF1 did not affect A4GALT transcript levels. Instead, protein pull-down experiments using P1 but not P2 oligonucleotide probes identified runt-related transcription factor 1 (RUNX1) by mass spectrometry. Furthermore, RUNX1 binds P1 alleles selectively, and knockdown of RUNX1 significantly decreased A4GALT transcription. These data indicate that RUNX1 regulates A4GALT and thereby the expression of clinically important glycosphingolipids implicated in blood group incompatibility and host–pathogen interactions.
Introduction
The P1 antigen was identified in 1927, shortly after the discovery of ABO.1 The P1PK blood group system includes 3 glycosphingolipid antigens, P1, Pk, and NOR, synthesized by different allelic variants of the A4GALT-encoded α1,4-galactosyltransferase.2,3 These histo-blood group antigens4,5 are important in transfusion medicine and also constitute receptors for microorganisms and toxins. Although crippling mutations in A4GALT abolish all 3 antigens,3 the molecular basis underpinning the common P1/P2 phenotypes has remained unknown. More A4GALT transcripts and higher Pk antigen levels were noted in P1-positive individuals,6,7 suggesting that P1/Pk expression may depend on transcription factor (TF) binding to regulatory regions of A4GALT. However, P1/P2-correlated variation was not found in the promoter,8,9 but in other noncoding sequences. We reported a single-nucleotide polymorphism (SNP) in exon 2a of A4GALT (rs8138197) that made P1/P2 genotyping possible and showed 99.5% and 100% concordance with P1 expression in 208 Swedish7 and 200 Taiwanese10 blood donors, respectively. Two SNPs (rs2143918/rs5751348), tightly linked to rs8138197 (Figure 1A), correlated fully and also exhibited transcription-inducing activity, indicating a role in A4GALT regulation.11 Interestingly, the rare In(Lu) phenotype that is due to Krüppel-like factor 1 (KLF1) haploinsufficiency results in lowered P1 expression.12 Based on these observations, we set out to unravel the molecular basis underlying the P1/P2 blood groups.
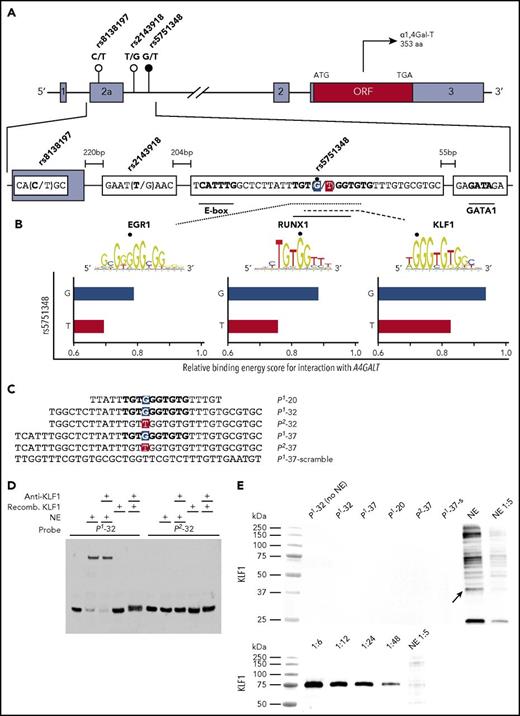
Presentation of the A4GALT gene, 3 candidate regulators, and gel shift assay showing no binding of KLF1 to the P1-specific rs5751348G sequence. (A) Schematic representation of A4GALT with the 3 SNPs suggested to determine the P1/P2 phenotypes. The main candidate SNP is marked with a black circle, with the others in white. Gray boxes depict noncoding exons with the open reading frame (ORF) in red and introns represented by horizontal black lines. In the lower, magnified section of exon 2a and the following intron, the P1-associated variants of each SNP and putative binding sites for selected TFs are indicated in boldface type. The main SNP (rs5751348) is highlighted in blue (P1 allele) and red (P2 allele). (B) EGR1, RUNX1, and KLF1 sequence logos showing binding preferences are adapted from the JASPAR database.25 The sequence logos for EGR1 and KLF1 are reversed to match the reading direction of A4GALT. Murine binding data are used for RUNX1 and KLF1 because data sets for the human TF are scarce (RUNX1) or not available (KLF1) in the database. For more data on RUNX1-binding requirements, see supplemental Figure 2. Bottom, 3 graphs highlighting the relative score of the binding energy for the 3 candidate TFs are displayed for both P1 and P2 alleles (G and T). The scale of the x-axis is centered around the default binding threshold of 0.8. (C) Oligonucleotides representing the sense strand in the probes used are shown with the nucleotide corresponding to rs5751348 with the P1 variant in blue and P2 in red. (D) EMSA shift and supershift reaction using P1/P2-32 probes, with nuclear extracts (NEs) from HEL cells or recombinant KLF1 and anti-KLF1. (E) Blotting with anti-KLF1 on protein pull-downs with P1 probes of various lengths, P2-37 probe and a scrambled version of the P1-37 probe. NE are shown as positive control for antibody binding and blotting of a serial dilution of recombinant KLF1 for anti-KLF1 to show appropriate detection by the antibody under the conditions used. Differences in molecular weights of KLF1 are due to an attached tag on the recombinant version of the protein.
Presentation of the A4GALT gene, 3 candidate regulators, and gel shift assay showing no binding of KLF1 to the P1-specific rs5751348G sequence. (A) Schematic representation of A4GALT with the 3 SNPs suggested to determine the P1/P2 phenotypes. The main candidate SNP is marked with a black circle, with the others in white. Gray boxes depict noncoding exons with the open reading frame (ORF) in red and introns represented by horizontal black lines. In the lower, magnified section of exon 2a and the following intron, the P1-associated variants of each SNP and putative binding sites for selected TFs are indicated in boldface type. The main SNP (rs5751348) is highlighted in blue (P1 allele) and red (P2 allele). (B) EGR1, RUNX1, and KLF1 sequence logos showing binding preferences are adapted from the JASPAR database.25 The sequence logos for EGR1 and KLF1 are reversed to match the reading direction of A4GALT. Murine binding data are used for RUNX1 and KLF1 because data sets for the human TF are scarce (RUNX1) or not available (KLF1) in the database. For more data on RUNX1-binding requirements, see supplemental Figure 2. Bottom, 3 graphs highlighting the relative score of the binding energy for the 3 candidate TFs are displayed for both P1 and P2 alleles (G and T). The scale of the x-axis is centered around the default binding threshold of 0.8. (C) Oligonucleotides representing the sense strand in the probes used are shown with the nucleotide corresponding to rs5751348 with the P1 variant in blue and P2 in red. (D) EMSA shift and supershift reaction using P1/P2-32 probes, with nuclear extracts (NEs) from HEL cells or recombinant KLF1 and anti-KLF1. (E) Blotting with anti-KLF1 on protein pull-downs with P1 probes of various lengths, P2-37 probe and a scrambled version of the P1-37 probe. NE are shown as positive control for antibody binding and blotting of a serial dilution of recombinant KLF1 for anti-KLF1 to show appropriate detection by the antibody under the conditions used. Differences in molecular weights of KLF1 are due to an attached tag on the recombinant version of the protein.
Study design
Identification of TF candidates
A 60-bp sequence centered around rs5751348 was examined for P1-specific TF-binding sites with bioinformatics tools (eg, MatInspector, v.8.0)13 and manually. Electrophoretic mobility-shift assay (EMSA) was performed with double-stranded probes and nuclear extracts (NEs) from human erythroleukemia (HEL) cells or recombinant KLF1. Anti-KLF1 was added for supershift. HEL-NE was incubated with different biotinylated probes. The probe–protein complexes were immobilized onto Dynabeads MyOne-Streptavidin-T1 (Thermo Fisher, Waltham, MA) and subjected to western blot or mass spectrometry. Probe sequences and further details on antibodies, proteins, and liquid chromatography-tandem mass spectrometry are shown in supplemental Tables 1 and 2 and supplemental Methods, available on the Blood Web site.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis/western blot
Samples were separated on AnykD Mini-Protean TGX Stainfree gels (Bio-Rad, Hercules, CA) followed by transfer to polyvinylidene difluoride membranes or Immun-Blot low-fluorescence polyvinylidene difluoride membranes (Bio-Rad). Membranes were incubated with anti-runt-related transcription factor 1 (RUNX1), anti-KLF1, or anti–early growth response factor 1 (EGR1). Recombinant KLF1 or EGR1 expressed in 293T/17 cells14 constituted positive controls. Chemiluminescence was detected using the ChemiDoc Touch Imaging System (Bio-Rad). (For further details, see supplemental Table 2 and supplemental Methods).
siRNA silencing of RUNX1 and KLF1
HEL and MEG-01 cells were used for small interfering (siRNA) silencing experiments. Cells were electroporated at a density of 7.5 × 106 cells/mL in RPMI-1640 (Thermo-Fisher), using 125 nM of each Silencer Select-siRNA (Thermo-Fisher). The siRNAs used were RUNX1-siRNA (s229351+s229352+s2460), KLF1-siRNA (s20962+s195062), negative control no.1 (cat. no. 4390843), and the positive control GAPDH-siRNA (s5573). Cells were electroporated at 280V/950 μF with square wave for 20 ms using the GenePulser MXcell Electroporation System (Bio-Rad). Following electroporation, cells were grown for 48 hours in 37°C, 5% CO2, in 4.9 mL RPMI-1640 with 10% fetal bovine serum (Thermo-Fisher).
Real-time quantitative polymerase chain reaction
RNA was extracted using RNeasy Plus Mini kit (Qiagen, Hilden, Germany), followed by DNase treatment and complementary DNA (cDNA) synthesis, using either TURBO DNA-free Kit and High-Capacity RNA-to-cDNA Kit, or Superscript IV Vilo according to the manufacturer’s instructions. Relative quantification of A4GALT (Hs00213726_m1), RUNX1 (Hs02558380_s1), KLF1 (Hs00610592_m1), and GAPDH (Hs02758991_g1 or Hs02786624_g1) was calculated using the ΔΔCT method with the endogenous control β-actin (HS01060665_g1) as calibrator.15 Results from Silencer Select Negative control no. 1, 1 μL of cDNA, was set to 1. Samples were run in technical triplicates using real-time polymerase chain reaction (QuantStudio 3) and analyzed using the QuantStudio Design-and-Analysis software, v1.4.1. All of these reagents, machinery, and software were from Thermo-Fisher. Statistical analysis was performed using the Mann-Whitney U test (IBM SPSS Statistics v23, IBM Corporation, Armonk, NY).
Results and discussion
Manual inspection and bioinformatic analysis of the sequence surrounding the P1/P2-differentiating G/T-SNP rs5751348 indicated potential binding sites for several TFs. Three candidate regulators were selected for further analysis based on their hematopoietic relevance and predicted decrease of their binding energy by substitution of G-P1 to T-P2 (Figure 1B). Candidates included KLF1, EGR1, and RUNX1. Although KLF1 is a well-known erythroid TF,16 EGR1 may promote myeloid17 or erythroid18 development and RUNX1 represses erythroid gene expression programs in favor of megakaryopoiesis.19 Because A4GALT is expressed also in other blood cells than erythrocytes4 and because the gene is transcribed early during hematopoiesis,20 all candidates were deemed interesting. KLF1 has been a long-standing A4GALT candidate regulator because of its involvement in the P1-weakening In(Lu) phenotype.12 We therefore first tested KLF1’s binding capacity to P1- and P2-specific probes of various lengths (Figure 1C). Although EMSA experiments showed clear shifts with P1 probes in the presence of nuclear extract (NE), no shifts were observed with recombinant KLF1 (Figure 1D), despite the latter being well detectable (Figure 1E). This negative finding was also supported by the absence of supershifts with anti-KLF1 (Figure 1D). To decipher the underlying reason for the apparently P1-specific EMSA shift with NE, magnetic beads were used to immobilize the biotinylated probes, capture the proteins bound, and subject them to liquid chromatography-tandem mass spectrometry. Strikingly, the top list of identified proteins (supplemental Table 3) revealed only 1 hematopoietic TF, RUNX1, with several unique peptides (supplemental Table 4) from different regions of the protein (Figure 2A). RUNX1 binding to P1-specific probes of 3 different lengths but not to P2-specific or scrambled probes was confirmed by western blot (Figure 2B). In addition, anti-RUNX1 displaced the shifted bands in an EMSA competition assay (Figure 2C). We found no evidence of EGR1 binding to the probes (supplemental Figure 1A). To investigate if any of the proteins identified by mass spectrometry that bind P1 but not P2 oligos might possibly interact with RUNX1, we performed STRING analysis (supplemental Methods and supplemental Table 5). Only low combined STRING scores or no scores at all were found for RUNX1 and selected other transcription factors. Notably, however, KLF1 and GATA1 had scores indicating possible interactions with RUNX1.
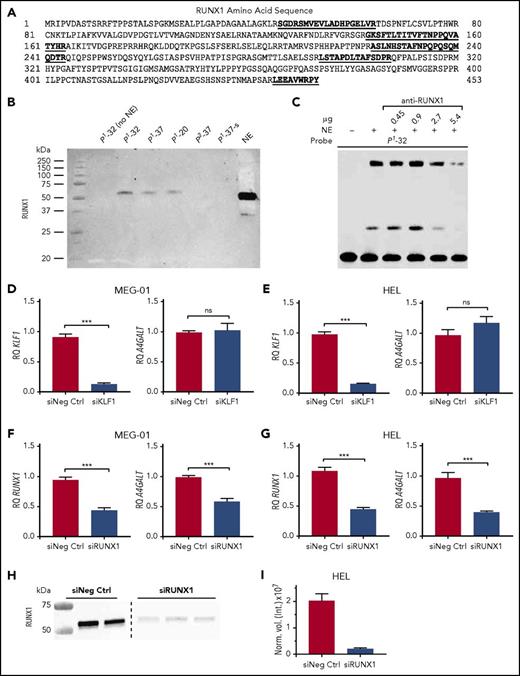
Identification of RUNX1 as the P1/P2-discriminating transcription factor binding to rs5751348G and its functional effect on A4GALT expression. (A) RUNX1 protein sequence, with all unique peptides identified by mass spectrometry marked in bold and underlined. (B) Blotting with anti-RUNX1 on protein pull-downs with P1 probes of various lengths, P2-37 probe, and a scrambled version of the P1-37 probe. NE are shown as positive control for antibody binding. (C) EMSA shift competition assay with P1-32 probe and increasing amounts of anti-RUNX1 to obtain blocking of shift. Relative quantification of KLF1 and A4GALT transcripts in siRNA-transfected MEG-01 (D) and HEL (E) cells. Relative quantification of RUNX1 and A4GALT transcripts in siRNA transfected MEG-01 (F) and HEL (G) cells. (D-G) Experiments were run in triplicate on 3 separate occasions. (H) Western blot of RUNX1 protein 48 hours after knockdown. The image displays 2 sections from the same blot, separated with a dashed line. (I) Quantification of the results in panel H, showing RUNX1 band intensity normalized to total protein content in lane. Error bars represent the standard error of the mean. *P < .05; **P < .01; ***P < .001 (Mann-Whitney U test). ns, not significant; RQ, relative quantity; siNeg Ctrl, negative control siRNA #1; siKLF1, 2 siRNAs targeting KLF1; siRUNX1, 2 or 3 siRNAs targeting RUNX1.
Identification of RUNX1 as the P1/P2-discriminating transcription factor binding to rs5751348G and its functional effect on A4GALT expression. (A) RUNX1 protein sequence, with all unique peptides identified by mass spectrometry marked in bold and underlined. (B) Blotting with anti-RUNX1 on protein pull-downs with P1 probes of various lengths, P2-37 probe, and a scrambled version of the P1-37 probe. NE are shown as positive control for antibody binding. (C) EMSA shift competition assay with P1-32 probe and increasing amounts of anti-RUNX1 to obtain blocking of shift. Relative quantification of KLF1 and A4GALT transcripts in siRNA-transfected MEG-01 (D) and HEL (E) cells. Relative quantification of RUNX1 and A4GALT transcripts in siRNA transfected MEG-01 (F) and HEL (G) cells. (D-G) Experiments were run in triplicate on 3 separate occasions. (H) Western blot of RUNX1 protein 48 hours after knockdown. The image displays 2 sections from the same blot, separated with a dashed line. (I) Quantification of the results in panel H, showing RUNX1 band intensity normalized to total protein content in lane. Error bars represent the standard error of the mean. *P < .05; **P < .01; ***P < .001 (Mann-Whitney U test). ns, not significant; RQ, relative quantity; siNeg Ctrl, negative control siRNA #1; siKLF1, 2 siRNAs targeting KLF1; siRUNX1, 2 or 3 siRNAs targeting RUNX1.
Finally, we asked if our findings would translate into functional consequences for A4GALT transcription. siRNA knockdown of KLF1 was efficient in 2 selected models (HEL and MEG-01 cells) but did not affect A4GALT transcript levels (Figure 2D-E). In contrast, siRNA knockdown of RUNX1 resulted in significant downregulation of A4GALT transcripts (Figure 2F-G). RUNX1 knockdown was most efficient at 24 hours for transcripts, but did not yet affect A4GALT levels (supplemental Figure 1B). Even though the RUNX1 transcript knockdown appeared less effective at 48 hours, RUNX1 protein was down to 10% of control (Figure 2H-I) at the point when A4GALT effect was observed (Figure 2F-G).
Taken together, these data suggest a role for RUNX1 in regulation of the A4GALT gene, its α1,4-galactosyltransferase, and the glycosphingolipids synthesized. Contrary to the hypothesis about KLF1 as a potential regulator of P1 expression, we identified RUNX1 in an unbiased way by a mass spectrometric approach following incubation of P1-specific probes with NE from erythroleukemic cells. Interestingly, RUNX1 is one of the major players in the TF network that represses erythroid differentiation. This is orchestrated through blocking of KLF1-dependent gene expression programs in the megakaryocytic lineage and is believed to be a crucial step at the megakaryocyte-erythroid progenitor stage.19 A4GALT transcripts can be detected as early as day 3 when growing primary bone marrow CD34+ cells toward the erythroid lineage.20 At this stage of culture, the cells have not yet become committed and only show early signs of erythroid differentiation. It is therefore possible that the P1/P2-differentiating effect of RUNX1 is initiated early and continued later, possibly by KLF1 or in complex with other TFs such as EGR1, even if we found no support for them in this study. Alternatively, KLF1 may have an altered binding affinity, which then blocks or disrupts the RUNX regulation. Even if P1 antigen is relatively restricted in its tissue distribution and mostly found on red blood cells, it has been described on mesothelioma and ovarian cancer cells.21,22 Pk, on the other hand, is found on several hematopoietic cell types, including red blood cells, as well as on other tissues,23 and is markedly increased in the P1 phenotype.7 Thus, quantitative regulation of A4GALT can be expected to involve also nonerythroid transcription factors and tissues.
In summary, this study unraveled a molecular mechanism underlying the expression of P1 antigen, a blood group discovered early but being one of the last to be explained. This new understanding could be of medical and biological value because A4GALT-dependent glycosphingolipid blood group antigens such as P1/Pk/NOR not only play vital roles in transfusion medicine, but are also known to serve as receptors for microbes and toxins. Their expression may therefore be a target for upregulation by pathogens as previously shown for certain other glycans.24
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Professor Urban Gullberg, Lund University, for kindly providing the EGR1 plasmid, anti-EGR1 (588), and 293T/17 cells. Support from the Swedish National Infrastructure for Biological Mass Spectrometry is gratefully acknowledged. Magnus Jöud, Lund University, is thanked for bioinformatic assistance.
This study was supported by grants from the Knut and Alice Wallenberg Foundation (2014.0312) and the Swedish Research Council (2014-71X-14251) (M.L.O.), Maggie Stephens’ Foundation (J.S.W.), and governmental Avtal om Läkarutbildning och Forskning (ALF) grants to the university health care in Region Skåne, Sweden (M.L.O.).
Authorship
Contribution: J.S.W., L.S., K.V., and S.K. performed experiments; J.S.W., L.S., K.V., Å.H., S.K., and M.L.O. interpreted and analyzed data; M.M. performed and interpreted bioinformatic analyses; J.S.W. and M.L.O. designed the research and wrote the paper; and all authors edited and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin L. Olsson, Division of Hematology and Transfusion Medicine, Department of Laboratory Medicine, Lund University, BMC C14, SE-22184 Lund, Sweden; e-mail: martin_l.olsson@med.lu.se.