TO THE EDITOR:
The B-cell receptor (BCR) signaling pathway plays a pivotal role in the pathogenesis of chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), and the use of inhibitors of the BCR pathway has demonstrated clinical activity in a variety of B-cell neoplasms.1,2 In recent clinical trials, CLL patients treated with the oral phosphatidylinositol 3-kinase δ (PI3K-δ) inhibitor idelalisib, showed significant progression-free survival and overall survival benefit.3,4 Duvelisib, a PI3K-δ/γ inhibitor with similar efficacy and toxicity profile, is currently being tested in clinical trials in various B-cell malignancies. However, relative to Bruton tyrosine kinase (BTK) inhibitors, the acquired resistance mechanisms of PI3K inhibitors (PI3Ki’s) and their long-term impacts on genetic alterations and clinical outcomes are largely unknown. A recent study demonstrated that PI3K-δ blockade could potentially induce genomic instability in the B-cells through activation of activation-induced cytidine deaminase (AID) that generates B-cell–specific random somatic mutations.5 Although high expression of AID was previously shown to have adverse prognostic impact in CLL and follicular lymphoma (FL) patients,6-8 the direct effect of AID-induced chromosome instability in the setting of PI3Ki treatment remains largely unknown. Here we report an unexpected clinical outcome: acute lymphoblastic leukemia (ALL) transformation in a CLL patient while he was receiving a PI3Ki, which requires attention in the use of these drugs.
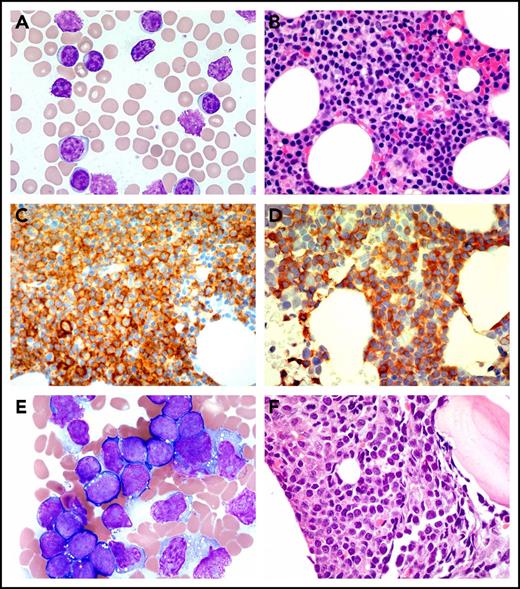
A 77-year-old man with history of CLL/SLL was referred to our cancer center for his newly diagnosed B-cell ALL. The patient initially developed left-sided axillary lymphadenopathy 4 years ago. Biopsies of lymph node and bone marrow (BM) revealed CD5-, CD19-, CD20-, and CD23-positive atypical lymphocytes on histochemistry and flow cytometry that were consistent with CLL/SLL (Figure 1A-D). No overexpression of CD38 or ZAP70 was noted. Further cytogenetics and molecular studies showed 11q23 deletion and TP53 mutations (TP53L265P, variant allele frequency [VAF] 2.0%; TP53G244D, VAF 4.0%) in the CLL cells. Because of his high-risk mutational profile, the patient was eligible for and enrolled in a phase 1 clinical trial of duvelisib, an oral PI3K-δ/γ inhibitor. The patient achieved partial response and remained on duvelisib 25 mg twice daily for 34 months until he developed progressive anemia and thrombocytopenia.
Peripheral blood and BM biopsy. (A) Peripheral blood smear (Wright stain, ×1000) and (B) BM core biopsy (Wright Giemsa, ×600) at the time of diagnosis of CLL. The neoplastic mature B-cells are small in size with clumped chromatin. (C) CD20 (immunoperoxidase, ×600) and (D) CD5 (immunoperoxidase, ×600) highlight the CLL cells. (E) BM aspirate (Wright Giemsa, ×1000) and (F) BM core biopsy (hematoxylin and eosin stain, ×600) at ALL diagnosis. The lymphoblasts are medium to large in size with fine chromatin, inconspicuous nucleoli, and scant basophilic cytoplasm with or without containing cytoplasmic vacuoles.
Peripheral blood and BM biopsy. (A) Peripheral blood smear (Wright stain, ×1000) and (B) BM core biopsy (Wright Giemsa, ×600) at the time of diagnosis of CLL. The neoplastic mature B-cells are small in size with clumped chromatin. (C) CD20 (immunoperoxidase, ×600) and (D) CD5 (immunoperoxidase, ×600) highlight the CLL cells. (E) BM aspirate (Wright Giemsa, ×1000) and (F) BM core biopsy (hematoxylin and eosin stain, ×600) at ALL diagnosis. The lymphoblasts are medium to large in size with fine chromatin, inconspicuous nucleoli, and scant basophilic cytoplasm with or without containing cytoplasmic vacuoles.
Complete blood count at that time showed white blood cells 9.7 × 109/L, hemoglobin 10.8 g/dL, and platelets 38 × 109/L. A flow cytometry on the peripheral blood showed CD5-negative, but CD34-, dim TdT-, and CD10-positive B cells. BM biopsy showed a hypercellular marrow with diffuse infiltrate by B-lymphoblastic leukemia cells that comprised 90% of the marrow cellularity (Figure 1E-F). Flow cytometry with BM sample confirmed the presence of a distinct lymphoblast population (94.3% of the total events) expressing CD10 (partial), CD19, CD20 (partial), CD34, CD38, cytoplasmic CD79a, nuclear TdT, and HLA-DR. Fluorescence in situ hybridization study was negative for del(11q23), del(13q14.3), del(17p13), trisomy 12, IgH-Bcl-1, and BCR-ABL1 rearrangement. Cytogenetics showed a normal male karyotype.
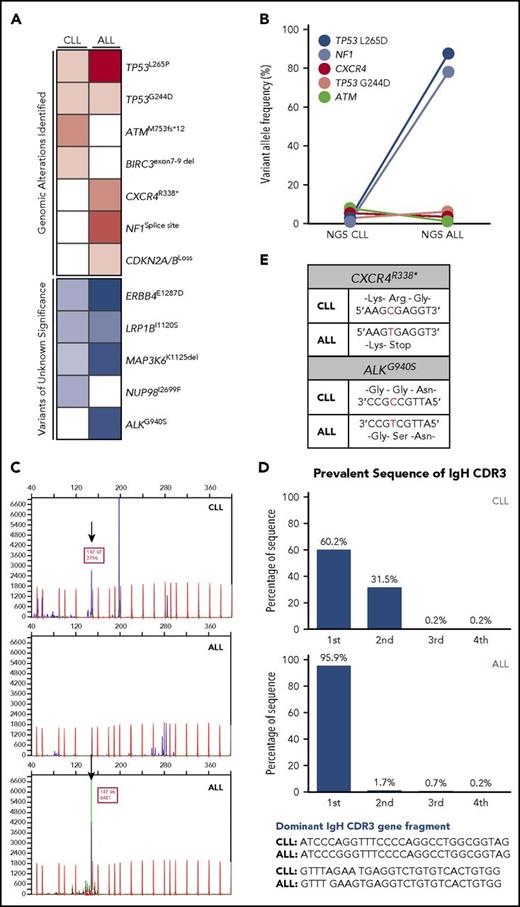
Next-generation sequencing (NGS) was performed with BM samples at the time of CLL and ALL diagnosis. Several gene mutations were identified in CLL BM sample including TP53L265P (2.0%), TP53G244D (4.0%), ATMM753fs*12 (7.4%), BIRC3 exons 7-9 deletion with additional variants of unknown significance including ERBB4E1287D (50.3%), LRP1BI1120S (49.6%), MAP3K6K1125del (45.4%), NUP98I2699F (45.4%), and mutational burden of 1 mutation/Mb (Figure 2A-B). In comparison, NGS of ALL BM sample showed an expansion of TP53L265P mutant clone (87.8%) as well as TP53G244D (2.0%), CXCR4R338* (6.8%), NF1 splice-site 5960_6006+118del165 (78.6%), and CDKN2A/B loss with additional variants of unknown significance including ALKG940S (60.7%), ERBB4E1287D (67.0%), LRP1BI1120S (64.0%), MAP3K6K1125del (63.2%), and tumor mutational burden of 3 mutations/Mb (Figure 2A-B), collectively suggesting a clonal expansion of TP53L265P-positive cells, which comprise the majority of ALL cells. Additional BCR gene rearrangements were performed to prove clonality and showed an identical size of clonal peak between CLL and ALL samples at molecular weight of 147.97 (Figure 2C). Subsequent sequencing of the IgH variable region gene demonstrated an identical sequence between these rearranged BCR gene segments (Figure 2D), confirming that both CLL and ALL cells share the common B-cell origin. In addition, the CXCR4 and ALK mutations were the consequence of C-to-T base substitution (Figure 2E), and 5′ preceding sequence of the CXCR4 mutation site matched with previously shown AID recognition motif [WRC; A/T(W), A/G(R)],5 indicating that ALL cells may have acquired CXCR4 mutation through AID activation by PI3K-δ blockade.
Analyses of clonality between CLL and ALL bone marrow samples. (A-B) NGS assessment was performed by Foundation One Heme, encompassing 406 genes and selected introns of 31 genes that are involved in the gene rearrangement and cancer related at the time of CLL and ALL diagnosis, respectively. DNA was extracted from BM samples (paraffin-embedded tissue) obtained at the diagnosis of CLL and ALL. Tumor mutational burden was measured by the number of somatic protein coding base substitution, insertion, and deletion mutations in the tumor specimen as part of NGS analyses. NGS showed an expansion of TP53L265D mutant clone. (C) For BCR gene rearrangement analysis, polymerase chain reaction (PCR) and capillary gel electrophoresis were adopted. DNA was extracted from paraffin-embedded BM cell clots and used in PCR amplification using Biomed-2 primers targeting all 3 immunoglobulin heavy chain (IgH) frameworks and immunoglobulin κ light chain (InVivoscribe Technologies) in fiver multiplex amplification tubes, each targeting conserved sequence in either framework (FR) 1, 2, or 3 of VH and DH in conjunction with a consensus (JH) primer in case of IgH. The primers for Vκ and Jκ, and Vκ and Kde are used for immunoglobulin κ light chain gene rearrangement. The PCR products were separated and detected by capillary gel electrophoresis on the ABI 3130x1. Blue and green peaks are PCR products amplified in CLL and ALL samples. Red peaks in the background stand for internal standard for molecular weight. BCR gene rearrangement analysis showed an identical monoclonal peak at molecular weight of 147.97 (black arrows). (D) Paraffin-embedded BM cell clots were obtained from the patient at diagnosis of CLL and B-ALL and sent to Adaptive Biotechnologies (Seattle, WA) for immunoglobulin heavy and light chain (VDJ, DJ, IgK) high-throughput sequencing using ClonoSeq assay to compare the sequence differences in clonal BCR gene rearrangements. The frequency of the most common 4 sequences (blue bars) identified on ClonoSeq analyses are presented. They are marked as first, second, third, and fourth in each CLL (upper) and ALL (bottom) sample. ClonoSeq confirmed identical IgH sequences of BCR gene rearrangement segments between CLL and ALL samples. (E) Sequencing of CXCR4 and ALK genes in CLL and ALL showed C-to-T mutation.
Analyses of clonality between CLL and ALL bone marrow samples. (A-B) NGS assessment was performed by Foundation One Heme, encompassing 406 genes and selected introns of 31 genes that are involved in the gene rearrangement and cancer related at the time of CLL and ALL diagnosis, respectively. DNA was extracted from BM samples (paraffin-embedded tissue) obtained at the diagnosis of CLL and ALL. Tumor mutational burden was measured by the number of somatic protein coding base substitution, insertion, and deletion mutations in the tumor specimen as part of NGS analyses. NGS showed an expansion of TP53L265D mutant clone. (C) For BCR gene rearrangement analysis, polymerase chain reaction (PCR) and capillary gel electrophoresis were adopted. DNA was extracted from paraffin-embedded BM cell clots and used in PCR amplification using Biomed-2 primers targeting all 3 immunoglobulin heavy chain (IgH) frameworks and immunoglobulin κ light chain (InVivoscribe Technologies) in fiver multiplex amplification tubes, each targeting conserved sequence in either framework (FR) 1, 2, or 3 of VH and DH in conjunction with a consensus (JH) primer in case of IgH. The primers for Vκ and Jκ, and Vκ and Kde are used for immunoglobulin κ light chain gene rearrangement. The PCR products were separated and detected by capillary gel electrophoresis on the ABI 3130x1. Blue and green peaks are PCR products amplified in CLL and ALL samples. Red peaks in the background stand for internal standard for molecular weight. BCR gene rearrangement analysis showed an identical monoclonal peak at molecular weight of 147.97 (black arrows). (D) Paraffin-embedded BM cell clots were obtained from the patient at diagnosis of CLL and B-ALL and sent to Adaptive Biotechnologies (Seattle, WA) for immunoglobulin heavy and light chain (VDJ, DJ, IgK) high-throughput sequencing using ClonoSeq assay to compare the sequence differences in clonal BCR gene rearrangements. The frequency of the most common 4 sequences (blue bars) identified on ClonoSeq analyses are presented. They are marked as first, second, third, and fourth in each CLL (upper) and ALL (bottom) sample. ClonoSeq confirmed identical IgH sequences of BCR gene rearrangement segments between CLL and ALL samples. (E) Sequencing of CXCR4 and ALK genes in CLL and ALL showed C-to-T mutation.
The patient was started on modified Dana-Farber Cancer Institute regimen9 and achieved morphologic complete remission after induction chemotherapy. Currently, he is on central nervous system phase treatment.
The BCR pathway has been shown to play an essential role in the pathogenesis of CLL/SLL, and inhibition of BCR pathway demonstrated clinical efficacy in a variety of B-cell neoplasms including CLL/SLL, FL, and mantle cell lymphoma.10-15 PI3K-δ is 1 of the 4 p110 catalytic isoforms that is highly expressed in lymphoid cells activating AKT/mTOR pathway,10-12,16,17 and phase 1/2 studies with idelalisib and duvelisib monotherapy showed promising results in relapsed/refractory low-grade B-cell lymphomas with overall response rate of 46% to 57%.18,19 In a pivotal phase 3 trial with relapsed CLL patients, combination of rituximab and idelalisib showed significant overall response and overall survival improvement compared with rituximab plus placebo group with acceptable side effect profiles,3 and idelalisib was approved by the US Food and Drug Administration for the treatment of relapsed CLL/SLL and FL. Currently, several PI3Ki’s are under clinical investigation, but long-term safety data are limited.
In comparison with ibrutinib, where the majority of early resistance arises from histologic transformation associated with TP53 mutation or CDKN2A/B loss vs late resistance from the BTK or PLCG2 genes mutations,20,21 the resistance mechanisms and the long-term effects of PI3K-δ blockades on the genetic alterations are largely unknown. A recent preclinical study demonstrated that PI3K-δ blockade induces marked elevation of AID, a B-cell–specific mutagenic enzyme, at the messenger RNA and protein levels, leading to genomic instability in a normal and a variety of B-cell malignant cells.5 Although high expression of AID was previously shown to have adverse prognostic impact in CLL and FL patients,6-8 the direct effect of AID-induced chromosome instability in the setting of PI3Ki treatment remains elusive given its short history of clinical use.
The CLL patient in our report developed a transformation to B-cell ALL while he was on duvelisib treatment. Our extensive laboratory analyses indicate that CLL and ALL cells are clonally related. NGS analyses showed an expansion of TP53L265P clone (VAF 87.8 from 2.0%) along with gain of new mutations in NF1 and CXCR4 genes and CDKNA/B loss at the time of ALL diagnosis. In addition, the mutational burden has increased from 1 up to 3 mutations/Mb following ALL transformation. Of note, nucleotide sequence preceding C-to-T mutation of CXCR4 gene in the ALL matched with AID recognition motif, which aligns with our hypothesis that AID may have induced these random mutations upon PI3K-δ blockade. BCR gene rearrangement and ClonoSeq analyses strongly support the clonality of CLL and ALL cells, although the similarity of light chains remains to be established. Collectively, these results suggest that PI3K-δ inhibition–induced AID could have generated random somatic mutations leading to genomic instability and subsequent CLL transformation into ALL.
We report the first case of transformed ALL from CLL under PI3Ki treatment. These therapies are given indefinitely, and patients are expected to be on medication for a long period of time until they develop progression of disease, loss of response, or intolerability. However, the effects of long-term use, specifically the impact on genomic alterations, are largely unknown. The precise identification of AID-dependent mutational signature is clinically important, and the long-term impact on the risk of Richter transformation or other aggressive forms of B-cell malignancy warrants further studies.
Authorship
Contribution: All authors were involved in patient care, participate in collecting and interpreting data, and writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Javier Pinilla-Ibarz, Malignant Hematology Department, H. Lee Moffitt Cancer Center & Research Institute, 12902 USF Magnolia Dr, Tampa, FL 33647; e-mail: javier.pinilla@moffitt.org.