Key Points
Xenografted ALL cells faithfully recapitulate CNS leukemia and are characterized by high expression of VEGF, mediating CNS entry of ALL cells.
VEGF captured by bevacizumab in vivo specifically reduces CNS leukemia, providing a novel strategy to target CNS involvement in ALL.
Abstract
In acute lymphoblastic leukemia (ALL), central nervous system (CNS) involvement is a major clinical concern. Despite nondetectable CNS leukemia in many cases, prophylactic CNS-directed conventional intrathecal chemotherapy is required for relapse-free survival, indicating subclinical CNS manifestation in most patients. However, CNS-directed therapy is associated with long-term sequelae, including neurocognitive deficits and secondary neoplasms. Therefore, molecular mechanisms and pathways mediating leukemia-cell entry into the CNS need to be understood to identify targets for prophylactic and therapeutic interventions and develop alternative CNS-directed treatment strategies. In this study, we analyzed leukemia-cell entry into the CNS using a primograft ALL mouse model. We found that primary ALL cells transplanted onto nonobese diabetic/severe combined immunodeficiency mice faithfully recapitulated clinical and pathological features of meningeal infiltration seen in patients with ALL. ALL cells that had entered the CNS and were infiltrating the meninges were characterized by high expression of vascular endothelial growth factor A (VEGF). Although cellular viability, growth, proliferation, and survival of ALL cells were found to be independent of VEGF, transendothelial migration through CNS microvascular endothelial cells was regulated by VEGF. The importance of VEGF produced by ALL cells in mediating leukemia-cell entry into the CNS and leptomeningeal infiltration was further demonstrated by specific reduction of CNS leukemia on in vivo VEGF capture by the anti-VEGF antibody bevacizumab. Thus, we identified a mechanism of ALL-cell entry into the CNS, which by targeting VEGF signaling may serve as a novel strategy to control CNS leukemia in patients, replacing conventional CNS-toxic treatment.
Introduction
Although pediatric acute lymphoblastic leukemia (ALL) has become a curable disease for many patients, involvement of the central nervous system (CNS) is still a major concern. By applying standard diagnostic procedures such as cytomorphology/cytospin analysis, CNS ALL is infrequently detected at initial diagnosis. However, CNS-directed therapy is essential for leukemia-free survival, even in patients without detectable CNS disease,1,2 indicating subclinical ALL manifestation at diagnosis in many patients. Clearly increased numbers of patients with CNS involvement would be detected, if more sensitive methods were applied.3-5 However, patients receiving CNS-directed intrathecal chemotherapy with or without cranial irradiation may experience neurocognitive deficits.6-12 To improve strategies to control CNS leukemia while decreasing the probability of secondary adverse effects and neurocognitive late sequelae, molecular mechanisms of leukemia-cell invasion into the CNS need to be better understood.
Several studies have shown links between different factors and CNS disease.13-18 However, none have demonstrated a role for leukemia-cell entry into the CNS. In T-cell ALL, chemokine signaling has been implicated in leukemia-cell survival, proliferation, and homing to the bone marrow (BM)19 as well as the CNS.20 Leukocyte entry into the CNS involves either migration of immune cells through the choroid plexus epithelium, transendothelial migration from postcapillary venules into the subarachnoid space and connected perivascular Virchow-Robin spaces, or direct migration from intraparenchymal microvessels into brain parenchyma, crossing the blood-brain barrier.21,22
CNS involvement in ALL has been described as a leptomeningeal disease, whereas parenchymal leukemia manifestation has been observed only in the late stages of the disease.23-25 Despite nondetectable CNS involvement by routine diagnostic methods in many cases, early autopsy studies have already shown that >50% of patients have submicroscopic CNS disease,25 highlighting the need for an appropriate model with which to study leukemia-cell entry into the CNS in vivo.
To characterize key molecular features of CNS disease in ALL, we used a nonobese diabetic (NOD)/severe combined immunodeficiency (SCID)/human ALL (huALL) xenotransplantation model26 and identified vascular endothelial growth factor A (VEGF) as a critical mediator of CNS disease in vivo, providing a rationale for anti-VEGF therapy in ALL.
Methods
NOD/SCID/huALL
Patient-derived xenograft samples (supplemental Table 1, available on the Blood Web site) were established by transplantation of patient ALL cells into 6-week-old female NOD/SCID mice (Charles River) as previously described.26 Patient samples were obtained after informed consent in accordance with the ethical review board of the institution (Ulm University); all animal experiments were approved and carried out in accordance with the appropriate authority (Regierungspräsidium Tübingen). When mice were moribund, they were euthanized by cervical dislocation, and organs were dissected. Single-cell suspensions were prepared by passing spleen tissue through nylon strainers as previously described26 or by flushing the BM cavities of both femora and tibiae of each animal. Meninges were dissected by carefully removing the upper cranial vault while detaching the meninges from the inner skull, cerebral cortex, and cranial base. Dissected meninges were mounted on a specimen slide for photography (Canon Powershot). Meningeal cell suspensions were prepared by passing the meningeal tissue through a nylon strainer, similar to the splenic cell preparation described earlier in this section. Leukemia loads were calculated as absolute numbers of human CD19+ cells analyzed by flow cytometry (BD LSR II flow cytometer). For in vivo treatment, mice receiving transplants were randomly assigned to either bevacizumab (10 mg/kg) or vehicle (phosphate-buffered saline) as 2 intraperitoneal injections per week, starting week 2 (X24) or 4 (X1, X1_1, X2) according to previously observed engraftment times.26 In sample X1, ALL engraftment was confirmed at the time of treatment start by detection of leukemia-specific T-cell receptor rearrangements (supplemental Table 2) identified at diagnosis (amplification of incomplete T-cell receptor Δ [Dδ2-Dδ3]) by quantitative polymerase chain reaction (qPCR; Applied Biosystems 7000) as previously described.27 Treatment was continued up to 13 weeks until mice showed clinical features of leukemia. For X1, 2 mice from each group underwent small-animal magnetic resonance imaging (MRI) and immunohistochemistry.
Gene-expression analyses
RNA was prepared by TRIzol (Invitrogen, Carlsbad, CA) extraction from leukemia samples (viable human CD19+ cells; median CNS, 98%; median BM, 97%). RNA was quality analyzed (Agilent 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA) and arrayed (Affymetrix Human Genome-U133 Plus 2.0; Affymetrix 3′-IVT Express Kit; Santa Clara, CA) according to manufacturer’s instructions. Gene-expression data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus28 (GEO series accession #GSE89710).
Genes of interest were validated by qPCR using the LightCycler Fast Start DNA Master SYBR Green I Kit following the manual (12239264001; Roche, Basel, Switzerland). Expression quantities were analyzed using the LightCycler 2.0 software followed by normalization to B2M, which was identified to be the most stably expressed housekeeping gene in the cohort applying the Normfinder algorithm.29 Gene-set enrichment was performed as described previously30 using R software (version 3.2.3; R Foundation, Vienna, Austria) to generate the gene matrix and comparing the expression profiles of CNS and BM samples using the C2cgp gene-set (Molecular Signatures Database) collection. Top-20 annotated gene sets are listed.
Cell culture and reagents
NALM-6 and KOPN-8 were purchased from DSMZ (ACC-128, ACC-552; Braunschweig, Germany), and CNS microvascular endothelial cells (ECs) bEnd.3 from ATCC (CRL-2299; Manassas, VA). 293FT cells were obtained from Invitrogen (R70007). Lines were maintained as described by the manufacturer. Cells were routinely checked for mycoplasma and authenticated by single-tandem repeat profiling.
For in vitro viability and proliferation assays, cells (500 000 cells/mL) were incubated with XTT (0.3 mg/mL; phenazine methosulfate 25 µM; 3 hours at 37°C), 100 μl isopropanol were added (30 minutes at room temperature), and absorbance (450 nm) was measured using a standard enzyme-linked immunosorbent assay plate reader. eFluor Cell Proliferation Dye 670 (0.5 µM; 65-0840-85; Affymetrix) was used following the manual. Viable cell numbers were analyzed by trypan blue exclusion. For analysis of apoptosis, cells were labeled with Nicoletti buffer (propidium iodide 50 µg/mL; 0.1% sodium citrate; 0.4% Triton-X 100; 1 hour at 4°C). Emission peaks were analyzed using a BD FACS Calibur flow cytometer. All experiments were performed in triplicate. For transwell assays, 100 000 bEnd.3 cells were seeded on 8-µm transwell inserts (353097; Corning, Tewksbury, MA) and cultured until reaching a confluent monolayer (4 days). 250 000 NALM-6 cells (10% fetal calf serum medium) were plated onto the inserts and allowed to migrate into the lower compartment (20% fetal calf serum medium; 48 hours). Viable cells were counted by trypan blue exclusion.
Immunohistochemistry and western blot
Immunohistochemistry was performed as described previously31 using anti-CD10 (56C6; Menarini, Florence, Italy) and anti-CD19 antibodies (LE-CD19; DAKO, Glostrup, Denmark). Pictures were taken with the Keyence (Neu-Isenburg, Germany) BZ-9000 microscope or Zeiss Axiophot (Oberkochen, Germany) using a 10× or 20× objective lens and a numerical aperture of 0.75 at room temperature. Pictures were analyzed using BZ-II Analyzer (Keyence) or Diskus (Königswinter, Germany), respectively.
For western blot analyses, protein was extracted as described previously.32 Protein (12 µg) was separated on 12% Bis-Tris gels, transferred onto membranes (NW00120BOX; iBLOT2 device IB21001; Thermofisher, Waltham, MA), and developed using a Protec optimax developer (Oberstenfeld, Germany).
Small-animal MRI
Narcotized mice (1.5% isoflurane; 80%/20% N2/O2) were imaged with a T2-weighed turbo spinecho technique for a maximum of 2 hours. MRI scans were analyzed using OsiriX (version 5.8; Pixmeo, Bernex, Siwtzerland).
Cloning
For stable VEGF knockdown, the Life Technologies BLOCK-iT™ Pol II miR RNAi Expression Vector Kit (K4935-00; Carlsbad, CA) was used following the manual. VEGF microRNA was designed using the Life Technologies RNAi Designer. Post-transduction, NALM-6 cells were sorted according to green fluorescent protein positivity. VEGF complementary DNA was cloned into pLenti6.3/V5-DEST following the manual of the pLenti6.3/V5-DEST Gateway Vector Kit (V533-06).
Positively transduced cells were selected (blasticidin 5 µg/mL). Knockdown and overexpression efficiency were analyzed by qPCR and enzyme-linked immunosorbent assay.
Statistical analyses
R and Prism 6.0 software (Graphpad Software, Inc., La Jolla, CA) were used to perform statistical analyses, considering P < .05 as significant and assuming equal variances. Gene-expression data were analyzed using the Bioconductor package for R (version 3.1.2.), applying the shrinkage t statistic, controlling for multiple testing, and maintaining a false-discovery rate < 0.05.33,34 Fluorescence-activated cell sorting data were analyzed using FlowJo software (version 8.7; Ashland, OR). Additional detailed methods can be found in the supplemental Data.
Results
Manifestation of CNS disease on xenotransplantation of primary BCP ALL cells
On transplantation of pediatric B-cell precursor (BCP) ALL cells into NOD/SCID mice, overt leukemia with high proportions of human ALL cells in peripheral blood, BM, and spleen was detected, as previously described.26 Interestingly, transplantation of some leukemias induced additional symptoms such as seizures and paraplegia, suggesting involvement of the CNS, whereas other samples did not induce these symptoms. To further investigate this phenotype of potential CNS involvement, we transplanted 15 leukemia samples (supplemental Table 1) into NOD/SCID mice. Nine of 15 samples led to these CNS symptoms. In addition to BM and spleen, enlarged meninges with a massive infiltration of high numbers of human CD19+ cells were identified in the mice showing CNS symptoms, reminiscent of meningeal leukemia-cell infiltration observed in patients with ALL with CNS disease (meningeosis leucaemica). In contrast to these CNS disease+ (CNS+) mice, no CNS disease (absence of CNS symptoms, no meningeal enlargement, and absent/very low meningeal infiltration together with high-level infiltration of BM and spleen) was observed in the other recipients (CNS disease− [CNS−]; Figure 1A-E; supplemental Figure 1A-D). Accordingly, massively enlarged meninges with infiltrations of human CD10+ ALL cells were detected by histology in CNS+ but not CNS− recipients (Figure 1F). Moreover, cerebral MRI of animals with manifest leukemia revealed thickened and enlarged meninges, corresponding to leukemia infiltration in CNS+ but not CNS− mice (Figure 1G-H).
Xenografting of primary ALL cells leads to meningeal infiltration, faithfully recapitulating CNS leukemia seen in patients. Recipient mice engrafted with xenograft X1 and X2 showed CNS disease (CNS+) with enlarged meninges (Canon Powershot macroscopic image of dissected meningeal tissues) (A) and infiltration of huCD19+ ALL cells in CNS, BM, and spleen (flow cytometry) (B) or no CNS disease (X10, X11) with no meningeal enlargement (C) and minimal CNS infiltration despite high-level BM and spleen infiltration (CNS−) (D). (E) Comparison of leukemia loads in CNS (P = .0018), BM (P = .5287), and spleen (P = .6889) in CNS+ (n = 9) and CNS− (n = 6) xenografts (bars indicate median; significance by 2-tailed Mann-Whitney U test). (F) Presence (X1; CNS+) or absence (X10; CNS−) of prominent meningeal infiltration of huCD10+ ALL cells in the leptomeningeal space outside the brain vessels (immunohistochemistry staining for huCD10; Zeiss Axiophot; size as indicated). MRI scans (T2-weighted transversal and coronal sections) showing prominent structures apical to the cerebral cortex (highlighted by dotted lines in 1 hemisphere) corresponding to enlarged and thickened meninges in CNS+ (G) but not in CNS− animals (H). (Note bright appearance of cerebrospinal fluid [CSF] in third or lateral ventricles in coronal sections). hu, human; mu, murine.
Xenografting of primary ALL cells leads to meningeal infiltration, faithfully recapitulating CNS leukemia seen in patients. Recipient mice engrafted with xenograft X1 and X2 showed CNS disease (CNS+) with enlarged meninges (Canon Powershot macroscopic image of dissected meningeal tissues) (A) and infiltration of huCD19+ ALL cells in CNS, BM, and spleen (flow cytometry) (B) or no CNS disease (X10, X11) with no meningeal enlargement (C) and minimal CNS infiltration despite high-level BM and spleen infiltration (CNS−) (D). (E) Comparison of leukemia loads in CNS (P = .0018), BM (P = .5287), and spleen (P = .6889) in CNS+ (n = 9) and CNS− (n = 6) xenografts (bars indicate median; significance by 2-tailed Mann-Whitney U test). (F) Presence (X1; CNS+) or absence (X10; CNS−) of prominent meningeal infiltration of huCD10+ ALL cells in the leptomeningeal space outside the brain vessels (immunohistochemistry staining for huCD10; Zeiss Axiophot; size as indicated). MRI scans (T2-weighted transversal and coronal sections) showing prominent structures apical to the cerebral cortex (highlighted by dotted lines in 1 hemisphere) corresponding to enlarged and thickened meninges in CNS+ (G) but not in CNS− animals (H). (Note bright appearance of cerebrospinal fluid [CSF] in third or lateral ventricles in coronal sections). hu, human; mu, murine.
We also addressed the stability of the CNS phenotype on serial and parallel transplantations. Spleen-derived ALL cells of 2 CNS+ (X1, X8) and 2 CNS− (X11, X12) leukemias were transplanted in up to 7 consecutive passages into up to 3 recipients in parallel. Importantly, CNS phenotypes were maintained on parallel and subsequent transplantations, and all animals showed high BM infiltration (supplemental Figure 1E-F). Thus, involvement and leukemia manifestation in the CNS seem to be leukemia-intrinsic features consistently recapitulated in our model system.
CNS ALL is characterized by high VEGF expression
To gain insight into the cellular profile of leukemia cells migrating to and residing in the CNS, sample pairs (n = 9) of ALL cells isolated from meningeal infiltrates or BM were investigated by whole human-genome expression analyses. Comparison of leukemia cells from both compartments identified 33 probe sets (27 genes) to be differentially regulated (shrinkage t statistic; false-discovery rate < 5%; fold change > +2 or < −2; Figure 2A; Table 1). The differentially expressed genes reflected a profile of hypoxia adaption (Figure 2B; supplemental Table 3). Interestingly, the gene coding for VEGF was determined to be highly upregulated in CNS-derived ALL cells (Figure 2A; Table 1). VEGF regulates, in addition to angiogenesis and EC growth, vascular permeability, transendothelial migration, and cellular survival.
CNS ALL cells are characterized by high VEGF expression and a profile associated with hypoxia adaptation. (A) Signature of 27 differentially regulated genes (33 probe sets; shrinkage t statistic; false-discovery rate [FDR] < 5%; fold change [FC] < −2 or FC > +2) comparing ALL sample pairs (n = 9) retrieved from CNS and BM identifying VEGF as a highly differentially regulated gene (red box; FC = 9.18; q = 9.68 × 10−13). Unsupervised cluster analysis according to this signature showing 2 clusters of CNS- and BM-derived ALL. (B) Gene-set enrichment analysis of CNS and BM signatures identified hypoxia-annotated gene sets (FDR q < 0.05). Enrichment score is shown as a green line, which reflects members of the annotated gene sets (black vertical lines) appearing along the gene list ranked from CNS (red) to BM (blue). (C) Differential VEGF expression (qPCR) in ALL sample pairs isolated from CNS and BM (BM CNS+; n = 9; P = .002) of CNS+ and BM samples derived from CNS− primografts (BM CNS−; n = 6; P = .0176). (D) qPCR analysis of VEGF expression in CNS and BM sample pairs of an independent cohort of CNS+ primografts (n = 8; P = .0009). Bars indicate median; significance by 2-tailed Mann-Whitney U test.
CNS ALL cells are characterized by high VEGF expression and a profile associated with hypoxia adaptation. (A) Signature of 27 differentially regulated genes (33 probe sets; shrinkage t statistic; false-discovery rate [FDR] < 5%; fold change [FC] < −2 or FC > +2) comparing ALL sample pairs (n = 9) retrieved from CNS and BM identifying VEGF as a highly differentially regulated gene (red box; FC = 9.18; q = 9.68 × 10−13). Unsupervised cluster analysis according to this signature showing 2 clusters of CNS- and BM-derived ALL. (B) Gene-set enrichment analysis of CNS and BM signatures identified hypoxia-annotated gene sets (FDR q < 0.05). Enrichment score is shown as a green line, which reflects members of the annotated gene sets (black vertical lines) appearing along the gene list ranked from CNS (red) to BM (blue). (C) Differential VEGF expression (qPCR) in ALL sample pairs isolated from CNS and BM (BM CNS+; n = 9; P = .002) of CNS+ and BM samples derived from CNS− primografts (BM CNS−; n = 6; P = .0176). (D) qPCR analysis of VEGF expression in CNS and BM sample pairs of an independent cohort of CNS+ primografts (n = 8; P = .0009). Bars indicate median; significance by 2-tailed Mann-Whitney U test.
CNS ALL gene signature
| Gene symbol . | Probe set . | FC . | q . |
|---|---|---|---|
| VEGF | 210512_s_at | 3.198580098 | 9.68E−13 |
| ADM | 202912_at | 2.297922281 | 0.000207549 |
| ABCA1 | 203504_s_at | 2.29578625 | 6.35E−06 |
| AK4 | 225342_at | 2.280186872 | 3.50E−07 |
| ABCA1 | 203505_at | 2.115501033 | 1.13E−05 |
| SPP1 | 209875_s_at | 2.056444216 | 6.23E−05 |
| ABCG1 | 204567_s_at | 1.965413943 | 0.000850829 |
| AK4 | 204348_s_at | 1.920807235 | 1.74E−06 |
| MAFF | 36711_at | 1.795093151 | 0.0124737 |
| NA | 242457_at | 1.637106403 | 0.029626065 |
| ATP1B1 | 201242_s_at | 1.588785175 | 1.72E−10 |
| MIR210HG | 230710_at | 1.582355482 | 9.68E−13 |
| ZIC1 | 206373_at | 1.431499478 | 9.68E−13 |
| MIR210HG | 236480_at | 1.426993584 | 9.68E−13 |
| CDH11 | 207173_x_at | 1.321728931 | 2.49E−12 |
| ANKRD37 | 227337_at | 1.313413756 | 9.68E−13 |
| NA | 236180_at | 1.302435902 | 9.68E−13 |
| P4HA1 | 207543_s_at | 1.274448098 | 9.68E−13 |
| PFKFB3 | 202464_s_at | 1.177839848 | 2.24E−05 |
| PFKFB4 | 228499_at | 1.1750446 | 5.24E−07 |
| AK4 | 204347_at | 1.166170764 | 7.92E−05 |
| MAF | 209348_s_at | 1.154728502 | 0.00016059 |
| HBA1 | 214414_x_at | 1.15260021 | 9.01E−06 |
| RBM24 | 235004_at | 1.151408134 | 0.035208142 |
| HSPA1A | 202581_at | 1.122039938 | 0.013751884 |
| FUT11 | 238551_at | 1.101206718 | 9.68E−13 |
| MYLIP | 223130_s_at | 1.092710897 | 0.000814121 |
| NA | 226347_at | 1.074786207 | 1.79E−12 |
| RORA | 226682_at | 1.066857298 | 5.41E−06 |
| FUT11 | 226348_at | 1.047230534 | 1.43E−09 |
| MYLIP | 228098_s_at | 1.032775799 | 0.01642794 |
| HK2 | 202934_at | 1.005172825 | 0.000107231 |
| COL3A1 | 215076_s_at | 1.00356502 | 9.68E−13 |
| Gene symbol . | Probe set . | FC . | q . |
|---|---|---|---|
| VEGF | 210512_s_at | 3.198580098 | 9.68E−13 |
| ADM | 202912_at | 2.297922281 | 0.000207549 |
| ABCA1 | 203504_s_at | 2.29578625 | 6.35E−06 |
| AK4 | 225342_at | 2.280186872 | 3.50E−07 |
| ABCA1 | 203505_at | 2.115501033 | 1.13E−05 |
| SPP1 | 209875_s_at | 2.056444216 | 6.23E−05 |
| ABCG1 | 204567_s_at | 1.965413943 | 0.000850829 |
| AK4 | 204348_s_at | 1.920807235 | 1.74E−06 |
| MAFF | 36711_at | 1.795093151 | 0.0124737 |
| NA | 242457_at | 1.637106403 | 0.029626065 |
| ATP1B1 | 201242_s_at | 1.588785175 | 1.72E−10 |
| MIR210HG | 230710_at | 1.582355482 | 9.68E−13 |
| ZIC1 | 206373_at | 1.431499478 | 9.68E−13 |
| MIR210HG | 236480_at | 1.426993584 | 9.68E−13 |
| CDH11 | 207173_x_at | 1.321728931 | 2.49E−12 |
| ANKRD37 | 227337_at | 1.313413756 | 9.68E−13 |
| NA | 236180_at | 1.302435902 | 9.68E−13 |
| P4HA1 | 207543_s_at | 1.274448098 | 9.68E−13 |
| PFKFB3 | 202464_s_at | 1.177839848 | 2.24E−05 |
| PFKFB4 | 228499_at | 1.1750446 | 5.24E−07 |
| AK4 | 204347_at | 1.166170764 | 7.92E−05 |
| MAF | 209348_s_at | 1.154728502 | 0.00016059 |
| HBA1 | 214414_x_at | 1.15260021 | 9.01E−06 |
| RBM24 | 235004_at | 1.151408134 | 0.035208142 |
| HSPA1A | 202581_at | 1.122039938 | 0.013751884 |
| FUT11 | 238551_at | 1.101206718 | 9.68E−13 |
| MYLIP | 223130_s_at | 1.092710897 | 0.000814121 |
| NA | 226347_at | 1.074786207 | 1.79E−12 |
| RORA | 226682_at | 1.066857298 | 5.41E−06 |
| FUT11 | 226348_at | 1.047230534 | 1.43E−09 |
| MYLIP | 228098_s_at | 1.032775799 | 0.01642794 |
| HK2 | 202934_at | 1.005172825 | 0.000107231 |
| COL3A1 | 215076_s_at | 1.00356502 | 9.68E−13 |
Differentially regulated genes comparing expression profiles of ALL primograft cells retrieved from CNS and BM (n = 9; 33 probe sets; 27 genes; false-discovery rate < 0.05; fold change [FC] > +2 or < −2; FC given as logarithm [base 2]).
Significantly upregulated VEGF expression was confirmed by qPCR in CNS-derived cells compared with corresponding BM samples and BM samples of CNS− cases (Figure 2C). In addition, it was reconfirmed in sample pairs of an independent set of CNS+ leukemias (Figure 2D; supplemental Table 1). Moreover, we verified detection of human VEGF using respective species-specific primer sets. Human VEGF was detected along with human B2M in ALL cells isolated from xenografted mice and the human cell line NALM-6, but not in CNS or BM specimens from mice not receiving transplants. Murine GAPDH was detected in all 4 mouse-derived specimens but not in human NALM-6 cells (supplemental Figure 2A).
VEGF is localized on chromosome 6p12 and comprises 8 exons. By alternative splicing, different isoforms can be transcribed, which have different receptor-binding capacities and differ in their pro- and antiangiogenic functions.35 By isoform-specific PCR, we only identified expression of VEGF165 and VEGF121 (supplemental Figure 2B-D). Both isoforms exhibit proangiogenic functions and bind to VEGF receptor 1 (VEGFR1) and VEGFR2.36 VEGFR1 was expressed in all primografts without a significant difference in CNS- or BM-derived cells of either CNS phenotype (Figure S2E, F), while VEGFR2 expression was not found in any of the leukemias (supplemental Figure 2G).
Previously, upregulated interleukin-15 (IL-15) transcript expression in BM samples of patients with ALL was reported to be associated with CNS involvement.13 However, no different IL-15 levels were found in CNS or BM samples of either phenotype (supplemental Figure 2H-I). Moreover, SPP1, which was also identified in our signature (Table 1), was recently described to be associated with CNS relapse.37 However, on validation in our cohort, upregulated SPP1 expression was only observed in some samples (supplemental Figure 2J-K).
NALM-6 and KOPN-8 induce a CNS+ phenotype
To provide a basis for in vitro molecular studies, we investigated the BCP ALL cell lines NALM-6 and KOPN-8, which both induced a CNS+ phenotype in vivo (supplemental Figure 3A-C). In both lines, VEGF transcripts (VEGF165 and VEGF121) were expressed at high levels, comparable to primografts (supplemental Figure 3D-E). Importantly, VEGF protein was detected in cell-culture supernatant of both lines (supplemental Figure 3G). As in xenografts, no VEGFR2 expression was detected, whereas both lines showed expression of VEGFR1 transcript and surface protein (supplemental Figure 3H-I).
VEGF-independent proliferation and survival of ALL cells
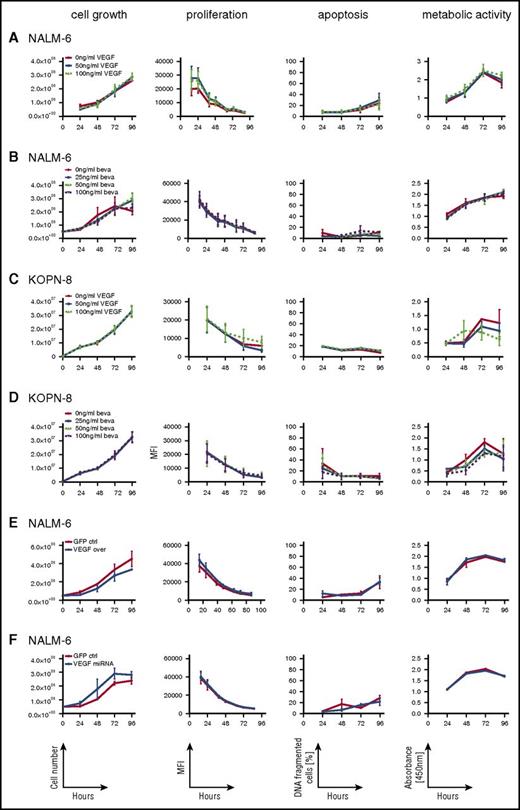
Given the expression of VEGF and VEGFR1 on ALL cells, we addressed whether VEGF is involved in autocrine regulation of leukemia growth and survival. NALM-6 and KOPN-8 were cultured in the presence of recombinant human VEGF or the VEGF-capturing antibody bevacizumab or were left untreated. Interestingly, neither the presence of up to 100 ng/mL of VEGF (Figure 3A,C) nor VEGF capture (Figure 3B,D) revealed any effect on cellular viability, survival, growth, or proliferation.
ALL cell growth, viability, proliferation, and apoptosis are not regulated by VEGF. Analysis of cellular growth (cell numbers), proliferation (eFluor; decrease of label intensity on cell division), apoptosis (DNA fragmentation), and cellular viability (XTT; quantification of substrate turnover) of NALM-6 and KOPN-8 cells cultured in the presence of recombinant VEGF (A,C) or the VEGF-capturing antibody bevacizumab (beva) (B,D) and of NALM-6 cells with stable VEGF overexpression (E) or VEGF downregulation (F), showing VEGF-independent growth, proliferation, apoptosis, and metabolic activity of ALL cells. Graphs show results of 3 independent experiments, each run in triplicate; mean and standard error of the mean are shown. ctrl, control; GFP, green fluorescent protein; MFI, mean fluorescent intensity; miRNA, microRNA.
ALL cell growth, viability, proliferation, and apoptosis are not regulated by VEGF. Analysis of cellular growth (cell numbers), proliferation (eFluor; decrease of label intensity on cell division), apoptosis (DNA fragmentation), and cellular viability (XTT; quantification of substrate turnover) of NALM-6 and KOPN-8 cells cultured in the presence of recombinant VEGF (A,C) or the VEGF-capturing antibody bevacizumab (beva) (B,D) and of NALM-6 cells with stable VEGF overexpression (E) or VEGF downregulation (F), showing VEGF-independent growth, proliferation, apoptosis, and metabolic activity of ALL cells. Graphs show results of 3 independent experiments, each run in triplicate; mean and standard error of the mean are shown. ctrl, control; GFP, green fluorescent protein; MFI, mean fluorescent intensity; miRNA, microRNA.
In addition, VEGF overexpression or microRNA-mediated downregulation in NALM-6 cells (supplemental Figure 4) did not affect growth, survival, or proliferative activity (Figure 3E-F). Thus, VEGF seems not to be involved in regulating overall survival or growth of ALL cells.
VEGF promotes transendothelial migration of ALL cells
CNS ALL is a leptomeningeal disease, with leukemia cells migrating from postcapillary venules through the endothelium and infiltrating the meningeal connective tissue and subarachnoid space. VEGF/VEGFR2 signaling contributes to vascular permeability involving downstream SRC/AKT kinase signaling in endothelial cells, thereby disrupting the endothelial barrier and leading to extravasation of tumor cells and metastasis.38-44
To analyze the effects of VEGF on ECs of the CNS, we used the murine CNS microvascular EC line bEnd.3 and assessed SRC and AKT activation. In contrast to leukemia cells, bEnd.3 cells express VEGFR2 (Figure 4A). In response to VEGF, bEnd.3 cells showed increased phosphorylation of SRC and AKT (Figure 4B), indicating VEGFR2 downstream signaling.
VEGF induces VEGFR2 signaling in and mediates ALL cell migration through CNS microvascular ECs. (A) VEGFR2 expression in CNS microvascular ECs (bEnd.3) but not NALM-6 BCP ALL cells. Western blot analysis; α-tubulin shows loading of equal amounts of protein. (B) Induction of VEGFR2 downstream signaling by AKT473− phosphorylation and SRC416− (western blot analysis) on VEGF exposure (100 ng/mL) at indicated increasing time points. (C) Migration of NALM-6 leukemia cells (red) through CNS microvascular ECs (blue). Increased transendothelial migration in the presence of VEGF (D) or of VEGF-overexpressing NALM-6 cells (E). Decreased migration on VEGF capture by bevacizumab (F) or of NALM-6 cells with VEGF downregulation (G). Data points represent numbers of migrated cells per well (n = 21, VEGF 0 ng/mL; n = 24, VEGF 50 ng/mL; n = 23, VEGF 100 ng/mL; n = 24, green fluorescent protein [GFP] control [ctrl] VEGF overexpression; n = 23, VEGF overexpression; n = 23, bevacizumab 0 ng/mL; n = 24, bevacizumab 25, 50, and 100 ng/mL; n = 24, GFP ctrl VEGF knockdown; n = 23, VEGF microRNA [miRNA]); bars indicate median values; significance by 2-tailed Mann-Whitney U test; P values as indicated in graphs.
VEGF induces VEGFR2 signaling in and mediates ALL cell migration through CNS microvascular ECs. (A) VEGFR2 expression in CNS microvascular ECs (bEnd.3) but not NALM-6 BCP ALL cells. Western blot analysis; α-tubulin shows loading of equal amounts of protein. (B) Induction of VEGFR2 downstream signaling by AKT473− phosphorylation and SRC416− (western blot analysis) on VEGF exposure (100 ng/mL) at indicated increasing time points. (C) Migration of NALM-6 leukemia cells (red) through CNS microvascular ECs (blue). Increased transendothelial migration in the presence of VEGF (D) or of VEGF-overexpressing NALM-6 cells (E). Decreased migration on VEGF capture by bevacizumab (F) or of NALM-6 cells with VEGF downregulation (G). Data points represent numbers of migrated cells per well (n = 21, VEGF 0 ng/mL; n = 24, VEGF 50 ng/mL; n = 23, VEGF 100 ng/mL; n = 24, green fluorescent protein [GFP] control [ctrl] VEGF overexpression; n = 23, VEGF overexpression; n = 23, bevacizumab 0 ng/mL; n = 24, bevacizumab 25, 50, and 100 ng/mL; n = 24, GFP ctrl VEGF knockdown; n = 23, VEGF microRNA [miRNA]); bars indicate median values; significance by 2-tailed Mann-Whitney U test; P values as indicated in graphs.
Moreover, we modeled transendothelial migration of ALL cells through bEnd.3 endothelial monolayers. In line with increased VEGFR2 downstream signaling, transendothelial migration of NALM-6 cells (Figure 4C) was clearly increased in the presence of VEGF, both on addition of recombinant VEGF and on VEGF overexpression (Figure 4D-E). Accordingly, VEGF capture by bevacizumab, as well as VEGF downregulation, led to a significant decrease of transendothelial leukemia-cell migration (Figure 4F-G).
VEGF capture inhibits CNS leukemia manifestation
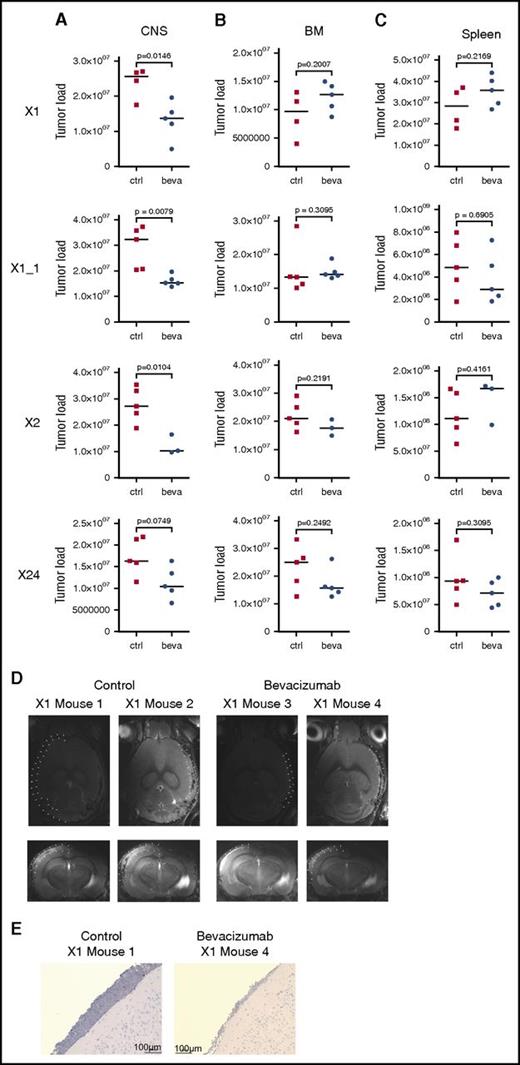
On the basis of these findings, we addressed the role of VEGF in mediating CNS involvement in vivo. Three individual primografts (X1, X2, and X24) all showing a CNS+ phenotype (Figure 1A-B; supplemental Figure 5A) and high VEGF expression in CNS-derived leukemia cells when compared with corresponding BM-derived samples (supplemental Figure 5B) were investigated in 4 experiments, including 1 replicate analysis (X1_1). Bevacizumab, which specifically binds to human but not to murine VEGF,45 was administered to CNS+ xenotransplant recipients. On onset of leukemia-related symptoms in control-treated animals, all mice were euthanized, and leukemia manifestation was quantified in CNS, BM, and spleen. Most interestingly, a clear reduction in CNS leukemia was seen in bevacizumab- as compared with control-treated animals in vivo (Figure 5A). However, no effect of leukemia reduction was observed in BM (Figure 5B) or spleen (Figure 5C) on bevacizumab treatment. In line with this observation, MRI performed after treatment of xenograft X1 revealed clearly reduced meningeal infiltrations in contrast to massively infiltrated meninges in control animals (Figure 5D). Accordingly, only low meningeal infiltration was observed in corresponding brain sections of bevacizumab-treated animals (Figure 5E). Expression analyses of VEGF in leukemia cells retrieved from CNS or BM of treated animals showed persistent high VEGF transcripts after bevacizumab treatment compared with control animals, both in CNS- and BM-derived cells (supplemental Figure 5C-F).
In vivo VEGF capture leads to compartment-specific reduction of leukemia load exclusively in the CNS. Leukemia load in CNS (A), BM (B), and spleen (C) of mice xenografted with 3 CNS+ primografts X1, X1_1 (replicate), X2, and X24 comparing control (ctrl)- and bevacizumab (beva)-treated recipients showing CNS compartment–specific leukemia reduction on VEGF capture (significantly lower CNS tumor load in X1, X1_1, and X2; trend toward lower CNS leukemia load in X24). X1: n = 4 for untreated, n = 5 for beva-treated; X1_1: n = 5 per group; X2: n = 5 for untreated, n = 3 for beva-treated; X24: n = 5 per group (bars indicate median values; significance by 2-tailed Mann-Whitney U test; P values as indicated in graphs). (D) Reduced meningeal ALL infiltration in post-treatment MRI analysis (transversal and coronal sections; meninges highlighted by dotted lines in 1 hemisphere) after bevacizumab compared with ctrl treatment. (E) Reduced meningeal infiltration of huCD19+ BCP ALL cells (immunohistochemistry staining for huCD19, BZ-9000; size as indicated).
In vivo VEGF capture leads to compartment-specific reduction of leukemia load exclusively in the CNS. Leukemia load in CNS (A), BM (B), and spleen (C) of mice xenografted with 3 CNS+ primografts X1, X1_1 (replicate), X2, and X24 comparing control (ctrl)- and bevacizumab (beva)-treated recipients showing CNS compartment–specific leukemia reduction on VEGF capture (significantly lower CNS tumor load in X1, X1_1, and X2; trend toward lower CNS leukemia load in X24). X1: n = 4 for untreated, n = 5 for beva-treated; X1_1: n = 5 per group; X2: n = 5 for untreated, n = 3 for beva-treated; X24: n = 5 per group (bars indicate median values; significance by 2-tailed Mann-Whitney U test; P values as indicated in graphs). (D) Reduced meningeal ALL infiltration in post-treatment MRI analysis (transversal and coronal sections; meninges highlighted by dotted lines in 1 hemisphere) after bevacizumab compared with ctrl treatment. (E) Reduced meningeal infiltration of huCD19+ BCP ALL cells (immunohistochemistry staining for huCD19, BZ-9000; size as indicated).
Thus, specific capture of human VEGF in vivo leads to reduced leukemia manifestation exclusively in the CNS compartment, indicating that leukemia manifestation in the CNS is mediated by human ALL-derived VEGF and, most importantly, can be reduced by VEGF inhibition.
Discussion
We observed that BCP ALL cells transplanted into NOD/SCID mice migrated into the CNS and were found to reside in the leptomeningeal compartments, faithfully recapitulating the clinical presentation of CNS leukemia observed in patients. Analysis of leukemia cells retrieved from the CNS revealed a gene-expression profile indicating hypoxia adaptation and high VEGF expression as compared with BM-derived cells. We found that transendothelial leukemia-cell migration was regulated by VEGF, and in vivo capture of VEGF showed a CNS compartment–specific effect with significantly decreased leukemia burden in CNS but not BM or spleen of leukemic mice, thus pointing to a novel mechanism of and strategy for controlling CNS leukemia.
Even in the 19th century, reports on autopsies of pediatric patients with leukemia described meningeal infiltration and invasion of the perivascular Virchow-Robin spaces by leukemia cells, thus characterizing CNS leukemia as a leptomeningeal disease.24,25 We also observed leptomeningeal leukemia infiltration in our NOD/SCID/huALL xenograft model on transplantation of patient-derived human ALL cells, mimicking the human pathology along with corresponding neurological symptoms and characteristic findings on MRI. Meningeal ALL in recipient mice had already been described in early transplantation experiments46,47 and in more recent studies using xenotransplantation of human ALL cells into immunodeficient mice.14,16,17,20,48 In all these studies, leukemia cells were inoculated into the BM niche, the supposed site of leukemia origination, or blood circulation, pointing to secondary spread and migration of ALL cells into the CNS.
However, CNS involvement observed on ALL transplantation seemed not to be associated with the CNS status of the corresponding patient,48 as also observed in our study, in which all samples were derived from patients without CNS involvement as assessed by cell counts or morphology, suggesting that ALL-cell entry into the CNS is a general leukemia-intrinsic feature and that subclinical CNS involvement remains undetected by standard cerebrospinal fluid (CSF) diagnostics in many patients with ALL.
Different studies have addressed cell entry into the CNS. IL-15 has been shown to be involved in CNS ALL, and high IL-15 mRNA expression in BM samples has been found to be associated with CNS involvement.13,14,17 In our analysis, IL-15 was not identified to be differentially regulated, in line with a study that found no difference in IL-15 expression in patient BM at diagnosis and isolated CNS relapse samples.37 That study also identified increased SPP1 expression in relapsed CNS ALL,37 as did we in our analysis. However, on validation, we did not observe significant differences in CNS- and BM-derived leukemias. In a similar model, CNS ALL was recently described on transplantation of the NALM-6 cell line and investigated by gene-expression analysis. Interestingly, VEGF was also found to be upregulated in CNS-derived NALM-6 cells.49 In t(1;19) ALL, Mer tyrosine kinase was shown to contribute to CNS leukemia on xenotransplantation,16 in line with the high risk for CNS relapse in this leukemia subgroup.50 In our study, all 3 t(1;19)+ leukemias induced CNS leukemia, as did the t(1;19)− leukemias. In T-cell ALL, the chemokine receptor CCR7 regulates leukemia-cell entry into the CNS, controlled by Notch1.20 In BCP ALL, however, contradictory findings have been reported. No association of chemokine or chemokine receptor expression with CNS leukemia was found in patient or xenograft samples, and receptor antagonization did not affect CNS engraftment,48,51 whereas another study reported ZAP70-mediated regulation of CCR7/CXCR4 and decreased CNS leukemia on CCR7 inhibition.52 In vitro studies have indicated involvement of adhesion molecules expressed on ALL cells in CNS migration, and coculture of meningeal and choroid plexus ECs and astrocytes with BCP-ALL lines conferred a chemotherapy-protective effect, suggesting that leukemia-cell adhesion might contribute to entry into and ALL-cell survival in the CNS.15,53,54
By direct comparison of pairs of CNS- and BM-derived ALL cells, we identified a CNS leukemia gene signature pointing to hypoxia adaptation of CNS-infiltrating ALL cells. Interestingly, ALL cells exposed to hypoxic conditions showed decreased sensitivity for prednisolone and methotrexate,55 2 important drugs used to control CNS ALL, thereby possibly indicating a mechanism of therapy escape. Among the identified genes, VEGF was the highest upregulated gene in CNS-derived cells. VEGF, initially cloned in HL-60 leukemia cells,56 controls neovascularization and endothelial growth, both in physiological and pathological situations, and is an important regulator of vascular permeability, transendothelial migration, and cellular survival.40,41,57,58
Increased angiogenesis and vascularization have been reported in BM specimens of pediatric ALL and BM of ALL-xenografted mice.58,59 Expression of VEGF and the isoforms VGEF165 and VEGF121 has been described in cells of different hematological malignancies, including pediatric ALL.60-63 However, we found VEGF to be highly upregulated only in CNS- but not BM-derived ALL cells. Most intriguingly, similar results were observed in patients with leukemia; significantly increased VEGF protein levels were identified in CSF specimens collected from patients with acute leukemia with CNS involvement as compared with low levels in corresponding serum samples. Moreover, CSF VEGF levels were significantly higher in patients with CNS+ as compared with CNS− leukemia or healthy controls, and high CSF VEGF levels were associated with inferior survival.64 In addition, monitoring VEGF protein levels in CSF in a patient with B-cell lymphoma revealed repeatedly increased VEGF levels at the time of CNS disease.65 Our data and these reports on high VEGF cytokine levels in the CSF of patients with CNS disease point to a substantial role of VEGF in mediating CNS leukemia. Moreover, by using a syngeneic virus-induced mouse leukemia model, meningeal leukemia-cell infiltration was observed, and most importantly, the leukemia cells were found to secrete high levels of VEGF,66 thereby further emphasizing the role of VEGF in mediating leukemia-cell entry into the CNS.
VEGF acts through binding to VEGFR1 and VEGFR2.36 Expression of both receptors has been described in cell lines and patient leukemia samples, with particularly high VEGFR1 expression in BM specimens from pediatric patients with ALL and expression of VEGFR2 mainly in myeloid leukemias.60,61,63,67-69 We found VEGFR1 expression in ALL primografts and cell lines, whereas VEGFR2 expression was not detected in any of the ALL cases. Despite VEGFR1 expression on ALL cells, we did not find evidence for involvement of VEGF in supporting leukemia-cell maintenance or growth. However, in vivo VEGF dependency was not analyzed, and additional analyses will reveal whether abrogation of VEGF signaling affects survival of leukemia cells in the CNS in vivo and thus whether this would be effective in treating manifest CNS leukemia.
VEGF also regulates the integrity of the endothelial barrier and contributes to extravasation of tumor cells and metastasis.40,41 Vascular permeability and EC survival is mediated through VEGFR2 and the downstream signaling kinases SRC and AKT.40-44 Importantly, in addition to activation of VEGFR2 downstream signaling, we observed increased or decreased transendothelial migration of ALL cells depending on the presence or absence of VEGF. Most importantly and in line with these findings, VEGF signaling has previously been implicated in CNS microvascular EC permeability.70,71 This further implies an important role of the VEGF/VEGFR2 axis in regulating ALL-cell migration through cerebral microvascular ECs, pointing to a role of anti-VEGF therapy in prevention of ALL-cell entry and CNS prophylaxis.
Thus, our study provides clear evidence that ALL-cell entry into the CNS is driven by VEGF, which is expressed and secreted by the leukemia cells and which binds to and activates VEGFR2 signaling, thereby mediating transendothelial leukemia-cell migration into the CNS. The importance of the concept of VEGF-driven CNS entry of ALL cells is further substantiated by the effect of in vivo VEGF capture by bevacizumab in leukemia-bearing mice receiving transplants of different CNS+ ALL primograft samples. Strikingly, we observed a CNS compartment–specific effect on bevacizumab treatment, with strongly reduced leukemia loads exclusively in the CNS. Because bevacizumab only captures human VEGF,45 the in vivo functional data on primary ALL cells not only underscore the role of VEGF in mediating CNS disease but also provide a novel therapeutic strategy to control CNS leukemia.
Presented in part at the 20th Congress of the European Hematology Association, Vienna, Austria, 11-14 June 2015, and the 57th Annual Meeting of the American Society of Hematology, Orlando, FL, 5-8 December 2015.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Volk and M. Herrmann for excellent technical assistance, Mike-Andrew Westhoff and Pamela Fischer-Posovszky for experimental support, and the Ulm University Sorting, Array, and Small Animal Imaging Core Facilities.
This work was supported by the International Graduate School in Molecular Medicine Ulm (V.M. and S.D.); German Research Foundation (KFO 167, K.-M.D.; SFB1074 B6, L.H.M. and K.-M.D.); Helmholtz Association (Preclinical Comprehensive Cancer Center) (K.-M.D.); Else Kröner-Fresenius Foundation, Ulm University Bausteinförderung (F.S.); and Förderkreis für Tumor und Leukämiekranke Kinder Ulm.
Authorship
Contribution: V.M., L.T., J.H., S.D., F.S., J.M.K., H.A.K., R.K., and T.F.E.B. performed research; V.M., L.T., J.H., S.D., F.S., J.M.K., H.A.K., R.K., T.F.E.B., G.t.K., K.-M.D., and L.H.M. analyzed and interpreted data; V.M., K.-M.D., and L.H.M. designed and conceptualized research and wrote the manuscript; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.T. is Laboratory of Oncohematology, Department of Women’s and Children’s Health, University of Padua, Padua, Italy.
The current affiliation for J.H. is Department of Internal Medicine III, Ulm University Medical Center, Ulm, Germany.
Correspondence: Lüder H. Meyer, Department of Pediatrics and Adolescent Medicine, Ulm University Medical Center, Eythstrasse 24, 89075 Ulm, Germany; e-mail: lueder-hinrich.meyer@uniklinik-ulm.de.
References
Author notes
K.-M.D. and L.H.M. share senior authorship.

![Figure 1. Xenografting of primary ALL cells leads to meningeal infiltration, faithfully recapitulating CNS leukemia seen in patients. Recipient mice engrafted with xenograft X1 and X2 showed CNS disease (CNS+) with enlarged meninges (Canon Powershot macroscopic image of dissected meningeal tissues) (A) and infiltration of huCD19+ ALL cells in CNS, BM, and spleen (flow cytometry) (B) or no CNS disease (X10, X11) with no meningeal enlargement (C) and minimal CNS infiltration despite high-level BM and spleen infiltration (CNS−) (D). (E) Comparison of leukemia loads in CNS (P = .0018), BM (P = .5287), and spleen (P = .6889) in CNS+ (n = 9) and CNS− (n = 6) xenografts (bars indicate median; significance by 2-tailed Mann-Whitney U test). (F) Presence (X1; CNS+) or absence (X10; CNS−) of prominent meningeal infiltration of huCD10+ ALL cells in the leptomeningeal space outside the brain vessels (immunohistochemistry staining for huCD10; Zeiss Axiophot; size as indicated). MRI scans (T2-weighted transversal and coronal sections) showing prominent structures apical to the cerebral cortex (highlighted by dotted lines in 1 hemisphere) corresponding to enlarged and thickened meninges in CNS+ (G) but not in CNS− animals (H). (Note bright appearance of cerebrospinal fluid [CSF] in third or lateral ventricles in coronal sections). hu, human; mu, murine.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/5/10.1182_blood-2017-03-769315/4/m_blood769315f1.jpeg?Expires=1769169771&Signature=uFhf5NtSJELHWuHOUKZ1PyvDYzSx-4hu~6Wz6c5MzKjjDtzB4P5rdFj6QOxSGFvbHhWwEpAYKVamSdZc2U92Ef5gnf-yPTqtn22aYA4ju2NSv3EGnzZHGf1knfMg1IEcrOVKoxYMME29B3lNSmvSYC8Pl1vYr4IrHQ0mVU8zv7hkl2m4pRT310l4vilYGVwHLOacnZYhN3kKe2ZcwZ6SvJjWSLyfQO0-4iumIoh3nWRxU2DlIjiUC-fZFqySBj8G9EOCyZ3Yi5OzDl4xYGIEeUKhjZYXCX-yNkTTmwObRy4bTaMr3sMl39etEs6zU8Bw31tROVh2asf1j09oyatvug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. CNS ALL cells are characterized by high VEGF expression and a profile associated with hypoxia adaptation. (A) Signature of 27 differentially regulated genes (33 probe sets; shrinkage t statistic; false-discovery rate [FDR] < 5%; fold change [FC] < −2 or FC > +2) comparing ALL sample pairs (n = 9) retrieved from CNS and BM identifying VEGF as a highly differentially regulated gene (red box; FC = 9.18; q = 9.68 × 10−13). Unsupervised cluster analysis according to this signature showing 2 clusters of CNS- and BM-derived ALL. (B) Gene-set enrichment analysis of CNS and BM signatures identified hypoxia-annotated gene sets (FDR q < 0.05). Enrichment score is shown as a green line, which reflects members of the annotated gene sets (black vertical lines) appearing along the gene list ranked from CNS (red) to BM (blue). (C) Differential VEGF expression (qPCR) in ALL sample pairs isolated from CNS and BM (BM CNS+; n = 9; P = .002) of CNS+ and BM samples derived from CNS− primografts (BM CNS−; n = 6; P = .0176). (D) qPCR analysis of VEGF expression in CNS and BM sample pairs of an independent cohort of CNS+ primografts (n = 8; P = .0009). Bars indicate median; significance by 2-tailed Mann-Whitney U test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/5/10.1182_blood-2017-03-769315/4/m_blood769315f2.jpeg?Expires=1769169771&Signature=Wbr9X2nFVypvYa87sh-YsZAToMhiScm5mdvf5Ts4Ksk39~-RknTTlz9BeAA0EtoKmgW690X4DB4bgzjuFCzuy2mKlYcRQFelvZhMkXAaePDZUFDBk3krv~G-xlDgWm3F2f3D5zq64M-AY~5D1VfYDxsY~L95mNc7tO4LpPc5f-s~yagr2-h2tHhuLBfAbFeqtl03I~EE3Td15ln~5w8zv2iiB~SW9KaFvBHEO3xgZNB7qK6EMkbur2s4S4oP113CFC6L7jG2UPIchCrdJ3KOBaQdGYp1gHJvpYDeMEKqLdNRVJb6~ULQ9oFsEXM7IfPBkotR77Le4jAf3BSuqQtDKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. VEGF induces VEGFR2 signaling in and mediates ALL cell migration through CNS microvascular ECs. (A) VEGFR2 expression in CNS microvascular ECs (bEnd.3) but not NALM-6 BCP ALL cells. Western blot analysis; α-tubulin shows loading of equal amounts of protein. (B) Induction of VEGFR2 downstream signaling by AKT473− phosphorylation and SRC416− (western blot analysis) on VEGF exposure (100 ng/mL) at indicated increasing time points. (C) Migration of NALM-6 leukemia cells (red) through CNS microvascular ECs (blue). Increased transendothelial migration in the presence of VEGF (D) or of VEGF-overexpressing NALM-6 cells (E). Decreased migration on VEGF capture by bevacizumab (F) or of NALM-6 cells with VEGF downregulation (G). Data points represent numbers of migrated cells per well (n = 21, VEGF 0 ng/mL; n = 24, VEGF 50 ng/mL; n = 23, VEGF 100 ng/mL; n = 24, green fluorescent protein [GFP] control [ctrl] VEGF overexpression; n = 23, VEGF overexpression; n = 23, bevacizumab 0 ng/mL; n = 24, bevacizumab 25, 50, and 100 ng/mL; n = 24, GFP ctrl VEGF knockdown; n = 23, VEGF microRNA [miRNA]); bars indicate median values; significance by 2-tailed Mann-Whitney U test; P values as indicated in graphs.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/5/10.1182_blood-2017-03-769315/4/m_blood769315f4.jpeg?Expires=1769169771&Signature=UCpW1QFCfGCyzrNV66~leI4GFPXXnxXCJIr9Mgog6M0OBbg2l9eXTakDVGPRFjw6JWZR77i90YmE0G0Bw7ACH5pdDwh3bjPVzaD8GEfBUJ0-QBocNuOlczy4kmcGLMyUCh-tQKKMS4XFZEFU~5-I4dy3RYJOFDEMZ~Yu350FMS5k~cSAYg95vK9iZUQxXtNdz9-ZTy5esahsuzx95gAfsAWV86~GAEdxfgqaiEE4~OtbsXyJagsg~mj0~k3lTYyWTKLmq810oEKFWEzplVED6A52iBFviIV1wenvKl3hup8cCKCBW88thMHnb-gDCZJq3hdI5aX5xdRp79wAnX3l5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal