Abstract
Philadelphia chromosome (Ph)-like acute lymphoblastic leukemia (ALL), also referred to as BCR-ABL1–like ALL, is a high-risk subset with a gene expression profile that shares significant overlap with that of Ph-positive (Ph+) ALL and is suggestive of activated kinase signaling. Although Ph+ ALL is defined by BCR-ABL1 fusion, Ph-like ALL cases contain a variety of genomic alterations that activate kinase and cytokine receptor signaling. These alterations can be grouped into major subclasses that include ABL-class fusions involving ABL1, ABL2, CSF1R, and PDGFRB that phenocopy BCR-ABL1 and alterations of CRLF2, JAK2, and EPOR that activate JAK/STAT signaling. Additional genomic alterations in Ph-like ALL activate other kinases, including BLNK, DGKH, FGFR1, IL2RB, LYN, NTRK3, PDGFRA, PTK2B, TYK2, and the RAS signaling pathway. Recent studies have helped to define the genomic landscape of Ph-like ALL and how it varies across the age spectrum, associated clinical features and outcomes, and genetic risk factors. Preclinical studies and anecdotal reports show that targeted inhibitors of relevant signaling pathways are active in specific Ph-like ALL subsets, and precision medicine trials have been initiated for this high-risk ALL subset.
Introduction
Acute lymphoblastic leukemia (ALL) is a constellation of genetic diseases driven by sentinel genetic alterations commonly derived from structural chromosome rearrangements, aneuploidy, and cooperative mutations in genes that encode for transcription factors regulating lymphoid development, tumor suppressors, proteins that regulate cell cycle progression, and epigenetic modifiers.1 Subtypes of ALL can be defined based on the nature of specific sentinel genetic aberrations, particularly chromosome translocations and interstitial rearrangements that create fusion genes encoding chimeric proteins or hijack gene promoters causing dysregulated oncogene expression.2
Philadelphia chromosome-positive (Ph+) ALL is defined by the t(9;22)(q34;q11) translocation that produces BCR-ABL1, a constitutively active tyrosine kinase. BCR-ABL1 fusion is present in essentially all cases of chronic myeloid leukemia and in ∼3% to 5% of pediatric ALL and 25% of adult ALL.3,4 Before the advent of tyrosine kinase inhibitor (TKI) therapy, Ph+ ALL was associated with very poor survival, which has been improved significantly by the early addition of imatinib or related TKIs to intensive chemotherapy regimens with or without hematopoietic stem cell transplantation (HSCT) in first complete remission.4-8
Initial identification of Philadelphia chromosome-like ALL
In 2009, Mullighan and colleagues from the Children’s Oncology Group (COG) and St. Jude Children’s Research Hospital (SJCRH) and den Boer and colleagues from the Netherlands described a new subtype of ALL, termed Philadelphia chromosome (Ph)-like (the term used herein) or BCR-ABL1–like ALL (the term we use for studies utilizing the Dutch definition), that had a gene expression profile highly similar to that of Ph+ ALL, a high frequency of deletions of IKZF1, which encodes the lymphoid transcription factor IKAROS, and other lymphoid transcription factor genes (eg, PAX5 and EBF1), and poor survival.9,10 Ph-like/BCR-ABL1–like ALL almost always occurs within the “B-other” subset lacking the sentinel B-ALL genetic alterations BCR-ABL1, ETV6-RUNX1, TCF3-PBX1, KMT2A (MLL) rearrangements, and high hyperdiploidy.10 Ph-like ALL is recognized as a provisional entity in the 2016 World Health Organization classification of myeloid neoplasms and acute leukemia,2 and publications from many groups have defined the myriad of genetic events that underlie Ph-like ALL, the frequency of Ph-like ALL in different age groups, and the associated clinical features and outcomes and have identified new opportunities for precision medicine therapies for this high-risk ALL subtype (Figure 1; Table 1).9-56
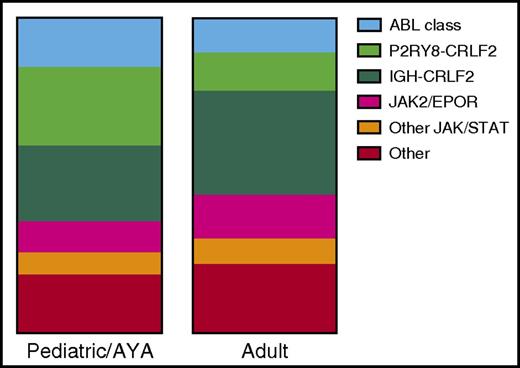
Incidence of Ph-like genetic alterations in children and adolescents and adults with B-lymphoblastic leukemia (B-ALL). Data are adapted from Reshmi et al51 (n = 284 patients) and Roberts et al52 (n = 194 patients). AYA, adolescents and young adults.
Kinase fusions identified in Ph-like ALL
| Kinase gene . | TKI . | Fusion partners identified to date, n . | 5′ Fusion partners . |
|---|---|---|---|
| ABL1 | Imatinib/dasatinib | 13 | CENPC, ETV6, FOXP1, LSM14A, NUP153, NUP214, RANBP2, RCSD1, SFPQ, SNX1, SNX2, SPTNA1, ZMIZ1 |
| ABL2 | Imatinib/dasatinib | 3 | PAG1, RCSD1, ZC3HAV1 |
| CSF1R | Imatinib/dasatinib | 3 | MEF2D, SSBP2, TBL1XR1 |
| PDGFRA | Imatinib/dasatinib | 1 | FIP1L1 |
| PDGFRB | Imatinib/dasatinib | 8 | ATF7IP, EBF1, ETV6, SNX29, SSBP2, TNIP1, ZEB2, ZMYND8 |
| LYN | Imatinib/dasatinib | 2 | GATAD2A, NCOR1 |
| CRLF2 | JAK2 inhibitor | 3 | CSF2RA, IGH, P2RY8 |
| JAK2 | JAK2 inhibitor | 21 | ATF7IP, BCR, EBF1, ETV6, GOLGA5, HMBOX1, OFD1, PAX5, PCM1, PPFIBP1, RFX3, SMU1, SNX29, SSBP2, STRN3, TERF2, TPR, USP25, ZBTB46, ZNF274, ZNF340 |
| EPOR | JAK2 inhibitor | 4 | IGH, IGK, LAIR1, THADA |
| TSLP | JAK2 inhibitor | 1 | IQGAP2 |
| TYK2 | TYK2 inhibitor | 3 | MYB, SMARCA4, ZNF340 |
| IL2RB | JAK1/JAK3 inhibitor | 1 | MYH9 |
| NTRK3 | TRK inhibitor | 1 | ETV6 |
| PTK2B | FAK inhibitor | 3 | KDM6A, STAG2, TMEM2 |
| FGFR1 | Ponatinib | 1 | BCR |
| FLT3 | FLT3 inhibitor | 1 | ZMYM2 |
| DGKH | 1 | ZFAND3 | |
| BLNK | 1 | DNTT | |
| CBL | 1 | KANK1 |
| Kinase gene . | TKI . | Fusion partners identified to date, n . | 5′ Fusion partners . |
|---|---|---|---|
| ABL1 | Imatinib/dasatinib | 13 | CENPC, ETV6, FOXP1, LSM14A, NUP153, NUP214, RANBP2, RCSD1, SFPQ, SNX1, SNX2, SPTNA1, ZMIZ1 |
| ABL2 | Imatinib/dasatinib | 3 | PAG1, RCSD1, ZC3HAV1 |
| CSF1R | Imatinib/dasatinib | 3 | MEF2D, SSBP2, TBL1XR1 |
| PDGFRA | Imatinib/dasatinib | 1 | FIP1L1 |
| PDGFRB | Imatinib/dasatinib | 8 | ATF7IP, EBF1, ETV6, SNX29, SSBP2, TNIP1, ZEB2, ZMYND8 |
| LYN | Imatinib/dasatinib | 2 | GATAD2A, NCOR1 |
| CRLF2 | JAK2 inhibitor | 3 | CSF2RA, IGH, P2RY8 |
| JAK2 | JAK2 inhibitor | 21 | ATF7IP, BCR, EBF1, ETV6, GOLGA5, HMBOX1, OFD1, PAX5, PCM1, PPFIBP1, RFX3, SMU1, SNX29, SSBP2, STRN3, TERF2, TPR, USP25, ZBTB46, ZNF274, ZNF340 |
| EPOR | JAK2 inhibitor | 4 | IGH, IGK, LAIR1, THADA |
| TSLP | JAK2 inhibitor | 1 | IQGAP2 |
| TYK2 | TYK2 inhibitor | 3 | MYB, SMARCA4, ZNF340 |
| IL2RB | JAK1/JAK3 inhibitor | 1 | MYH9 |
| NTRK3 | TRK inhibitor | 1 | ETV6 |
| PTK2B | FAK inhibitor | 3 | KDM6A, STAG2, TMEM2 |
| FGFR1 | Ponatinib | 1 | BCR |
| FLT3 | FLT3 inhibitor | 1 | ZMYM2 |
| DGKH | 1 | ZFAND3 | |
| BLNK | 1 | DNTT | |
| CBL | 1 | KANK1 |
Definitions of Ph-like and BCR-ABL1–like ALL differ among groups
Ph-like/BCR-ABL1–like ALL is defined by a gene expression profile and not by a single unifying sentinel molecular aberration. There is significant overlap between the Ph-like and BCR-ABL1–like ALL signatures, and most cases are concordant, but some cases are discordantly defined as Ph-like by COG/SJCRH and BCR-ABL1–like by the Dutch group.39
Den Boer and colleagues originally used the top probe sets predictive of BCR-ABL1 fusion in an earlier SJCRH study57 to develop a 110–probe set classifier that was highly accurate in classifying ALL cases into 6 subsets, one of which was B-ALL with BCR-ABL1.10 Hierarchical clustering of probe set expression patterns identified a subset of B-other ALL patients that resembled the BCR-ABL1+ cases. These BCR-ABL1–like patients had an increased relapse risk and inferior disease-free survival. The Dutch group has used hierarchical clustering with these 110 probe sets to define BCR-ABL1–like ALL in a number of subsequent studies.31,38,39,48
The COG/SJCRH group has taken 2 parallel approaches to define and identify Ph-like ALL. Mullighan and colleagues identified a subset of high risk BCR-ABL1–negative B-ALL cases that had IKZF1 deletions or point mutations and poor outcome.9 Because IKZF1 deletions are also very common in Ph+ ALL,58 they used Affymetrix U133 Plus 2.0 gene expression profiles to define the signature of BCR-ABL1–negative ALL patients associated with poor outcome (called Ph-like ALL), and a signature that defined BCR-ABL1+ cases. Gene-set enrichment analysis showed a highly significant similarity between these 2 expression signatures. Parallel unsupervised hierarchical clustering analysis of gene expression profiles using the same discovery set of patient samples identified 8 different cluster groups.15 One of these, the R8 group, was characterized by high-level expression of a set of genes that included the cytokine receptor-like factor 2 gene (CRLF2), frequent deletions of IKZF1 and other lymphoid transcription factor genes, Hispanic ethnicity, and poor survival and was highly overlapping with the Ph-like group.9,15 The COG/SJCRH group has subsequently used 2 methods to identify Ph-like patients. The first performs Predictive Analysis of Microarrays59 on array-based gene expression or RNA-sequencing (RNAseq) data to identify a signature associated with BCR-ABL1 that defines Ph-like cases.27,34 The second method identified subsets of 8 or 15 genes that can be analyzed rapidly via 384-well low-density microarray (LDA) cards to define Ph-like ALL51,52 and is currently used in COG clinical trials to identify patients in real-time for additional genomic screening.
There are several fundamental differences between analytic approaches that likely help explain why there is some discordance between Ph-like and BCR-ABL1–like ALL. The COG/SJCRH models have been refined over time and optimized to specifically identify Ph-like cases with defined genetic lesions, whereas the Dutch group has used a standardized set of genes over time to define BCR-ABL1–like ALL. The Dutch BCR-ABL1–like ALL signature includes 110 probe sets, and the COG/SJCRH Ph-like ALL signature includes 255 probe sets, and only 9 of these (recognizing 7 genes) are shared. None of these 9 probe sets are included in the 8-gene LDA Ph-like ALL signature used by the COG.39,51 CRLF2 is not included in the Dutch BCR-ABL1 signature or the COG/SJCRH Predictive Analysis of Microarrays signature, but is one of the core genes that define the COG R8 group and is one of the top genes in the COG 8-gene LDA used to identify Ph-like ALL.10,15,27,48,51 However, it is the underlying genomic alterations that are potentially clinically actionable, and not the Ph-like/BCR-ABL1–like status.
Sentinel molecular abnormalities in Ph-like ALL
Rearrangements of CRLF2 and JAK mutations
CRLF2 monomers pair with the interleukin-7 receptor-α (IL7R-α) subunit to form the heterodimeric thymic stromal lymphopoietin receptor (TSLPR) implicated in early B-cell development.60 About half of Ph-like ALL and 15% to 20% of BCR-ABL1–like ALL cases have 1 of 2 different genomic rearrangements involving CRLF2 (CRLF2-Rs), located on the pseudoautosomal region 1 (PAR1) of chromosomes Xp22/Yp11, resulting in high-level CRLF2 cell surface expression.10,11,13,14,17,38,39,48,61 Focal interstitial PAR1 deletion joins CRLF2 to the first noncoding exon of the adjacent P2RY8 gene, leading to the production of P2RY8-CRLF2 transcripts (Figure 2).11,13,14,17,61 Translocations, often cryptic, also join CRLF2 to the chromosome 14q32 immunoglobulin heavy chain gene IGH.13,14,17,61 Fusion of CRLF2 to another PAR1 gene, CSF2RA, has also been described.43 A recurrent activating CRLF2 F232C point mutation has also been reported, typically in association with CRLF2-Rs.17 A minority of CRLF2-Rs, particularly P2RY8-CRLF2, occur in cases that do not express the Ph-like ALL signature.14,51 In contrast, the frequency of high CRLF2 expression and P2RY8-CRLF2 fusion and JAK mutations is very similar in B-other cases with and without the BCR-ABL1–like phenotype.39,48 The different CRLF2-R frequencies may be due to the age and ethnic composition of patients studied, the algorithm used to define Ph-like/BCR-ABL1–like ALL, and whether or not CRLF2 fluorescence in situ hybridization (FISH) is performed routinely.
CRLF2 rearrangements in Ph-like ALL. (A) Interstitial deletion of the pseudoautosomal region (PAR1) of chromosomes X or Y places CRLF2 under control of the noncoding P2RY8 or CSF2RA promoters (dotted bar) to drive fusion transcript expression (thin arrows) and protein (thick arrows) translation (thin arrows). (B) IGH-CRLF2 fusion results from t(X;14) or t(Y;14). IGH is noncoding in the fusions (dotted bar). Both rearrangements result in CRLF2 overexpression and increased CRLF2 protein expression detectable by flow cytometry. (C) Rare CRLF2 point mutations lead to CRLF2 homodimerization and constitutive kinase signaling.
CRLF2 rearrangements in Ph-like ALL. (A) Interstitial deletion of the pseudoautosomal region (PAR1) of chromosomes X or Y places CRLF2 under control of the noncoding P2RY8 or CSF2RA promoters (dotted bar) to drive fusion transcript expression (thin arrows) and protein (thick arrows) translation (thin arrows). (B) IGH-CRLF2 fusion results from t(X;14) or t(Y;14). IGH is noncoding in the fusions (dotted bar). Both rearrangements result in CRLF2 overexpression and increased CRLF2 protein expression detectable by flow cytometry. (C) Rare CRLF2 point mutations lead to CRLF2 homodimerization and constitutive kinase signaling.
Activating JAK2 or JAK1 point mutations occur in about half of CRLF2-R ALL cases.11-14,17,61 The most common mutation is R683G in the JAK2 pseudokinase domain. JAK1 mutations are less common, but functionally analogous, and include JAK1 V658F, the homolog of the JAK2 V617F mutation seen in adult myeloproliferative neoplasms.62 CRLF2 overexpression and JAK2 mutations collaborate in transformation assays in vitro and lead to constitutive activation of JAK-STAT signaling (Figure 3).11,17,24,61,63
Schema of activated kinase signaling in Ph-like ALL. Kinase fusions and other alterations in Ph-like ALL activate oncogenic signal transduction and may be targetable by specific kinase inhibitors and other therapeutic agents.
Schema of activated kinase signaling in Ph-like ALL. Kinase fusions and other alterations in Ph-like ALL activate oncogenic signal transduction and may be targetable by specific kinase inhibitors and other therapeutic agents.
CRLF2-R and JAK2/JAK1 mutations are greatly enriched in children with Down syndrome–associated B-ALL (DS-ALL).11,13,61,64-66 Overall, 30% to 50% of children with DS-ALL have CRLF2-R compared with 5% to 10% of ALL patients without Down syndrome.11,13,14,51,53,61,65,66 Children with DS-ALL and CRFL2-R are more likely to have P2RY8-CRLF2 than IGH-CRLF2 and have a higher frequency of concomitant JAK mutations than those without Down syndrome.51,53 CRLF2-R DS-ALL cases also commonly have concomitant loss-of-function mutations in the deubiquitinase USP9X, which appears to suppress activated JAK2 signaling in these leukemias.67
P2RY8-CRLF2 fusion has been reported as significantly more common than IGH-CRLF2 in children, with ratios of 2:1 to 5:1 in some studies, but the relative incidence is affected by the type of analyses performed and patient characteristics.14,22,23,25,34,51,53,55 P2RY8-CRLF2 fusion is readily identified via reverse transcription polymerase chain reaction (RT-PCR), whereas the identification of IGH-CRLF2 requires FISH. Many published studies have not performed comprehensive CRLF2 FISH, likely underestimating the true incidence of IGH-CRLF2. Clinical features and ethnicity are also associated with the type of CRLF2-R. Patients with P2RY8-CRLF2 are younger (median age, 4 years) than those with IGH-CRLF2 (median age, 14 years) and also have lower white blood cell (WBC) counts, so they are less likely to be classified as National Cancer Institute high-risk (NCI HR; age ≥10 years or initial WBC count ≥50 000/μL; 38% vs 77%).53 Three studies of adolescents and adults with B-ALL found that IGH-CRLF2 was 2 to 4 times as common as P2RY8-CRLF2, emphasizing the variation in Ph-like ALL subtypes across the age spectrum.25,50,52
COG studies have consistently found that IGH-CRLF2 is associated with self-declared Hispanic ethnicity.14 Reshmi et al studied children and adolescents with NCI HR ALL or standard risk ALL with positive minimal residual disease (MRD) at the end of induction therapy using both P2RY8-CRLF2 polymerase chain reaction and IGH-CRLF2 FISH and reported a 1.03:1 ratio of P2RY8-CRLF2:IGH-CRLF2 alterations. In this analysis, 59% of children with IGH-CRLF2 were Hispanic vs 30.2% of those with P2RY8-CRLF2 and 27.6% of the studied B-ALL patients.51
JAK2 fusions and truncating rearrangements of the erythropoietin receptor
Constitutive JAK-STAT signaling is associated with 2 other recurrent alterations in Ph-like ALL (Figures 3 and 4). Translocations or interstitial deletions generating JAK2 fusion genes and JAK2 chimeric proteins occur in ∼5% of pediatric Ph-like ALL cases and more frequently in young adults.27,34,45,48,50-52 The amino terminus of JAK2 fusion proteins is encoded by the partner gene fused in-frame to the carboxyl terminal portion of JAK2, including the kinase (TK) domain with or without the upstream pseudokinase domain.16,27,34,40,42,43,45,48,51,52,68 The expression of JAK2 fusion proteins in vitro results in STAT5 activation and abrogates growth factor dependence in murine lymphoid growth factor–dependent cell lines (eg, Ba/F3), and treatment with ruxolitinib or other JAK2 inhibitors reverses both phenotypes.24,27,34,42,63,69,70
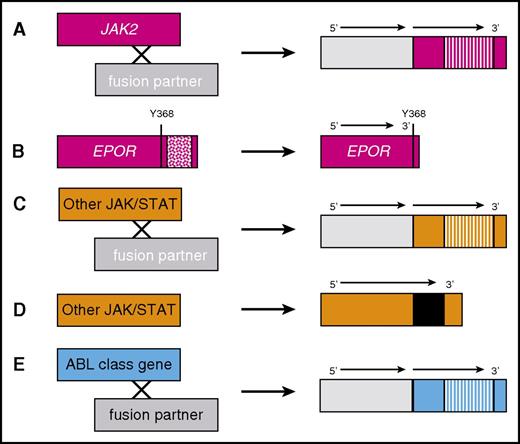
ABL-class kinase fusions and other JAK pathway alterations in Ph-like ALL. (A) JAK2 fusions are caused by translocations or interstitial deletions involving various 5′ fusion partners that lead to constitutive activation of the JAK2 3′ gene tyrosine kinase domain (striped bars). (B) Rearrangements of the EPOR gene with accompanying frameshift or stop codon mutations lead to overexpression of a truncated EPOR protein that lacks negative regulatory tyrosine (Y) residues (patterned box). Other JAK/STAT alterations (TYK2, IL7R, and TSLP) are caused by (C) translocations or (D) insertions, deletions, or missense mutations (black bar). (E) ABL-class (eg, ABL1, ABL2, CSF1R, and PDGFRB) kinase fusions occur via similar mechanisms as in panel A.
ABL-class kinase fusions and other JAK pathway alterations in Ph-like ALL. (A) JAK2 fusions are caused by translocations or interstitial deletions involving various 5′ fusion partners that lead to constitutive activation of the JAK2 3′ gene tyrosine kinase domain (striped bars). (B) Rearrangements of the EPOR gene with accompanying frameshift or stop codon mutations lead to overexpression of a truncated EPOR protein that lacks negative regulatory tyrosine (Y) residues (patterned box). Other JAK/STAT alterations (TYK2, IL7R, and TSLP) are caused by (C) translocations or (D) insertions, deletions, or missense mutations (black bar). (E) ABL-class (eg, ABL1, ABL2, CSF1R, and PDGFRB) kinase fusions occur via similar mechanisms as in panel A.
Rearrangements of the erythropoietin receptor (EPOR-R) involving ≥4 partner genes (IGH, IGK, LAIR1, and THADA; Table 1) are a recurrent alteration in Ph-like ALL, with an overall frequency of ∼1% among B-ALL cases.27,34,44,51,71 EPOR-Rs cause overexpression of C-terminal truncated EPOR proteins that have lost negative regulatory domains (Figure 3), resulting in JAK-STAT signaling activation (Figure 4) and in vitro sensitivity to ruxolitinib.44 Germ line mutations in EPOR produce analogous truncated EPOR proteins and cause autosomal dominant benign erythrocytosis.72
Other JAK/STAT-activating lesions
Uncommon deletions of SH2B3 (encoding LNK, a negative regulator of JAKs) and IL7RA insertions/deletions occur in Ph-like ALL and lead to activated signaling that is sensitive to JAK inhibition (Figure 3).21,24,73 Rare cases with other structural mutations have been reported and may also be sensitive to JAK inhibitors (Table 1).34
ABL-class fusions
The ABL-class fusion genes and chimeric proteins are structurally analogous to the JAK2 fusions described above and include rearrangements (translocations or intrachromosomal deletions/inversions) that target ABL1 itself, ABL2, CSF1R, or PDGFRB.27,34,39,45-48,51,52,74 A single case of an adult with FIP1L1-PDGFRA Ph-like ALL has recently been described, which may expand the spectrum of ABL-class fusions.52 ABL-class fusions occur in ∼3% to 5% of pediatric ALL, quite similar to the incidence of Ph+ ALL, whereas ∼2% to 3% of adult ALL cases have ABL-class fusions.34,51,52 The encoded chimeras include the carboxyl terminal portion of the ABL-class protein with its TK domain intact joined in-frame to the amino terminal portion of the partner protein (Figure 4). Just like BCR-ABL1, the ABL-class fusion proteins abrogate IL3 dependence in Ba/F3 cells and are similarly sensitive to imatinib and dasatinib that target their structurally homologous TK domains (and presumably other TKIs that target BCR-ABL1, although these have not been tested extensively to date).27,34 Imatinib and dasatinib also show potent clinical activity in primary patient samples and patient-derived xenograft (PDX) models of Ph-like ALL with ABL-class fusions.27,34
Uncommon fusions involving kinase genes
Other rare fusions involving kinase genes have been reported in Ph-like ALL (Table 1), including ETV6-NTRK3 fusion,34,51 which also occurs in infantile fibrosarcoma and secretory breast cancer.75,76 TRK inhibitors are being tested in clinical trials (eg, NCT02576431) with highly promising results across a spectrum of adult and pediatric solid tumors sharing TRK fusions.77 Other rare kinase fusions in Ph-like ALL involve BLNK, DGKH, FGFR1, IL2RB, LYN, PTK2B, TYK2, and RAS pathway genes.34,43,45,51,52,54,78 The functional properties of rare kinase fusions have generally not been validated, and only FGFR1 has a Food and Drug Administration–approved kinase inhibitor, ponatinib, which was developed for use against the BCR-ABL1 T315I gatekeeper mutation.
Other genomic alterations in Ph-like ALL
Although they usually accompany other genomic rearrangements, mutations and/or deletions in IKZF1, PAX5, and EBF1 have been reported in isolation in Ph-like ALL.34 Isolated mutations in the Ras pathway also occur in a small number of Ph-like ALL cases, but have been reported in conjunction with CRLF2 overexpression with and without JAK alterations.34,50
Germ line genomic factors linked to Ph-like ALL
Several North American studies have reported that Ph-like ALL is more common in persons of self-declared Hispanic/Latino ancestry.14,50,51 Genome-wide association studies have shown that the risk of developing Ph-like ALL is elevated in persons with specific GATA3 polymorphisms, and this link is particularly strong for Ph-like ALL with CRLF2-R.79,80 GATA3 risk alleles occur significantly more frequently in individuals with high Native American genetic ancestry and US Hispanics than in those of European descent.79
Clinical features and outcomes of Ph-like ALL in children and adults
The incidence of Ph-like ALL increases across the age spectrum and has been associated with inferior clinical outcomes in most studies. One small SJCRH study showed equivalent outcomes for Ph-like and non–Ph-like ALL patients, but 15% of Ph-like ALL patients underwent HSCT in first remission due to poor MRD response.35 Peak prevalence appears to occur in adolescence and young adulthood, although additional studies have recently confirmed that the Ph-like subtype is common in adults >40 years of age with B-ALL.49-52,80,81 The relative frequency of different sentinel genetic alterations differs between pediatric and adult ALL (Figure 1), with adults having a higher frequency of JAK2 fusions, EPOR-R, and, among CRFL2-R cases, more IGH-CRLF2 than P2RY8-CRLF2 compared with children.51,52 Ph-like ALL is associated with other high-risk clinical features besides age, including elevated WBC count, high rates of end-induction MRD, and increased risk of treatment failure and relapse.9,10,15,30,31,34,35,45,48-51 The inferior survival for Ph-like ALL occurs regardless of the underlying genomic alteration, and survival is particularly poor for Ph-like patients with elevated end-of-induction MRD. Some patients with Ph-like ALL experience overt induction failure, particularly those with PDGFRB-R.29,32,34,46
Precision medicine opportunities for treatment of Ph-like ALL
Because of the poor outcome associated with Ph-like ALL and the nature of the underlying sentinel genetic aberrations, there is great interest in using targeted therapies or precision medicine approaches. Although the Ph-like gene expression signature is useful diagnostically, it is not a therapeutic target. The prognostic significance and potential for therapeutic targeting of the IKZF1 deletion remains unclear given its association to date with both unfavorable (Ph+, Ph-like)9,10,23,28,31,48,58,82-84 and favorable (DUX4/ERG-dysregulated)85-87 B-ALL subtypes. ALL-associated IKZF1 alterations have been linked with upregulation of multiple genes involved in cellular proliferation and chemoresistance.88 One study found that IKZF1-deleted ALL patients benefited from periodic vincristine/steroid pulses during maintenance therapy,83 prompting the Dutch Childhood Oncology Group to extend maintenance therapy and to include vincristine and steroid pulses for patients with IKZF1 deletions (www.trialregister.nl; NTR3379). The benefit of maintenance vincristine/steroid pulses for patients with IKZF1-deleted ALL was not confirmed by other groups.89 Furthermore, the COG uses many more maintenance vincristine/steroid pulses than other groups and still finds IKZF1 deletion to be an adverse prognostic factor. Preclinical studies have demonstrated the therapeutic potential of retinoic acid compounds and FAK inhibitors in IKZF1-deleted ALL models, but these approaches have not yet been tested clinically.90,91 Deletions involving other B-lymphoid transcription factor genes, such as EBF1 and PAX5, are also recurrent alterations in Ph-like ALL, but strategies to target these lesions therapeutically have not yet been developed. The greatest opportunity for precision medicine approaches in Ph-like ALL is to use therapies targeted at the underlying sentinel molecular lesions, many of which are likely to be drivers of leukemogenesis. Different diagnostic approaches to identify these lesions have been used. As more cases are analyzed, more fusion partners and splice variants of known fusions have been identified for the ABL-class, JAK2, and rarer kinase genes (Table 1), emphasizing the need to use unbiased sequencing technologies, such as RNAseq.34,51,52 Analysis that relies only on RT-PCR for known fusions will invariably miss some, and perhaps many, targetable alterations.
Opportunities for treatment of patients with ABL-class fusions
Experimentally, ABL-class fusions phenocopy BCR-ABL1 and are similarly sensitive to imatinib and dasatinib.27,34 There is significant anecdotal evidence that imatinib and dasatinib can induce remissions and clear MRD in patients with Ph-like ALL and ABL-class fusions that have responded poorly to chemotherapy.20,29,32,34,46 Based on these factors, there is strong interest in testing TKI therapy in this high-risk ALL subtype. Clinical experience in Ph+ ALL has established that imatinib or dasatinib can be added safely to combination chemotherapy regimens.3,4,6-8 The real challenge is how to identify these patients in real-time given the variety of ABL-class fusions that currently includes ≥13 ABL1, 3 ABL2, 3 CSF1R, and 7 PDGFRB fusions (Table 1), which can have different breakpoints producing different fusion transcripts and with new fusion partners identified on a regular basis. A second major challenge is the rarity of this ALL subtype. Because ABL-class fusions occur in <5% of ALL cases, there are only a few hundred patients diagnosed in the United States each year, including both adults and children. Implementation of randomized trials of chemotherapy with and without added TKI therapy thus is logistically difficult and creates significant challenges in study design and interpretation, although several single-arm trials have been initiated (Table 2).
Current clinical trials of TKI-based therapies in children and adults with Ph-like ALL
| Ph-like ALL alterations . | Kinase inhibitor . | Disease status . | Age, y . | Clinical trial . |
|---|---|---|---|---|
| ABL class | Dasatinib | Newly diagnosed | 1-30 | NCT01406756 (COG AALL1131) |
| ABL class | Dasatinib | Newly diagnosed | 1-18 | NCT03117751 (SJCRH Total XVII) |
| ABL class | Dasatinib | Relapsed | ≥10 | NCT02420717 (MDACC) |
| CRLF2/JAK pathway | Ruxolitinib | Newly diagnosed | 1-21 | NCT02723994 (COG AALL1521) |
| CRLF2/JAK pathway | Ruxolitinib | Newly diagnosed | 1-18 | NCT03117751 (SJCRH Total XVII) |
| CRLF2/JAK pathway | Ruxolitinib | Relapsed | ≥10 | NCT02420717 (MDACC) |
| Ph-like ALL alterations . | Kinase inhibitor . | Disease status . | Age, y . | Clinical trial . |
|---|---|---|---|---|
| ABL class | Dasatinib | Newly diagnosed | 1-30 | NCT01406756 (COG AALL1131) |
| ABL class | Dasatinib | Newly diagnosed | 1-18 | NCT03117751 (SJCRH Total XVII) |
| ABL class | Dasatinib | Relapsed | ≥10 | NCT02420717 (MDACC) |
| CRLF2/JAK pathway | Ruxolitinib | Newly diagnosed | 1-21 | NCT02723994 (COG AALL1521) |
| CRLF2/JAK pathway | Ruxolitinib | Newly diagnosed | 1-18 | NCT03117751 (SJCRH Total XVII) |
| CRLF2/JAK pathway | Ruxolitinib | Relapsed | ≥10 | NCT02420717 (MDACC) |
There are several diagnostic strategies that could be used to identify ABL-class fusions. The COG developed a complex diagnostic strategy that first performs LDA to identify patients with Ph-like ALL enrolled in their AALL1131 trial (NCT02883049) and then uses a series of multiplex RT-PCR assays, FISH, and DNA sequencing to identify the underlying genomic aberration (Figure 5).51 Patients with Ph-like ALL and ABL-class fusions have dasatinib added to chemotherapy starting with the second month of treatment. Accrual to this treatment stratum began in late 2016. Because there is limited knowledge about the outcome of Ph-like ALL patients with ABL-class fusions that have good risk prognostic factors, such as young age, lower WBC count, or excellent early MRD response, the dasatinib arm of COG AALL1131 is limited to children with NCI HR ALL.
Current Ph-like ALL genetic testing algorithm used by the Children's Oncology Group. Diagnostic leukemia cells from children with high risk B-ALL are first screened for the Ph-like ALL gene expression signature using an 8-gene low density microarray (LDA) that includes CRLF2 as one of the 8 assessed genes.51 Specimens with the Ph-like ALL signature that lack BCR-ABL1 fusion (Ph+ ALL) undergo additional genetic testing. Those with high CRLF2 expression are assessed for P2RY8-CRLF2 and IGH-CRLF2 rearrangements by RT-PCR and FISH, respectively, and for JAK1, JAK2 and IL7R mutations by PCR. Ph-like ALL specimens without CRLF2 overexpression undergo multiplex RT-PCR fusion testing to detect ABL-class, JAK2, and EPOR rearrangements. Not all Ph-like fusions will be detected by this algorithm. Complete assessment may require alternative assays for identification, such as RNA-sequencing or unbiased fusion testing capable of identifying new 5′ partners. HR B-ALL, high-risk B-ALL.
Current Ph-like ALL genetic testing algorithm used by the Children's Oncology Group. Diagnostic leukemia cells from children with high risk B-ALL are first screened for the Ph-like ALL gene expression signature using an 8-gene low density microarray (LDA) that includes CRLF2 as one of the 8 assessed genes.51 Specimens with the Ph-like ALL signature that lack BCR-ABL1 fusion (Ph+ ALL) undergo additional genetic testing. Those with high CRLF2 expression are assessed for P2RY8-CRLF2 and IGH-CRLF2 rearrangements by RT-PCR and FISH, respectively, and for JAK1, JAK2 and IL7R mutations by PCR. Ph-like ALL specimens without CRLF2 overexpression undergo multiplex RT-PCR fusion testing to detect ABL-class, JAK2, and EPOR rearrangements. Not all Ph-like fusions will be detected by this algorithm. Complete assessment may require alternative assays for identification, such as RNA-sequencing or unbiased fusion testing capable of identifying new 5′ partners. HR B-ALL, high-risk B-ALL.
The SJCRH group is performing RNAseq in all patients enrolled in their Total XVII study, which opened for enrollment in early 2017 (NCT03117751), with results available before the end of the first month of therapy. Patients found to have ABL-class fusions have dasatinib added to the chemotherapy backbone. Most other groups do not yet have the ability to perform real-time RNAseq on all newly diagnosed ALL patients enrolled in their trials.
In 2015, the MD Anderson Cancer Center (MDACC) opened a phase 2 trial of dasatinib in combination with multiagent chemotherapy in older children and adults with relapsed/refractory Ph-like ALL and ABL-class fusions (NCT02420717). Eligible patients are initially treated with up to 3 weeks of dasatinib monotherapy with the addition of multiagent chemotherapy for those patients with suboptimal response to single-agent therapy. The primary outcome measurement is complete response rate after 6 weeks of therapy.
Opportunities for treatment of patients with JAK/STAT pathway lesions
Preclinical studies have demonstrated constitutive activation of kinase signaling networks in vitro in subsets of Ph-like ALL harboring JAK pathway alterations, including CRLF2-R with or without JAK mutations, JAK2 fusions, EPOR-R, and other alterations (Figure 3).12,13,17,24,27,34,44,63,92 Additional studies have reported in vivo activity of various JAK inhibitors in PDX models of JAK pathway–mutant Ph-like ALL, providing rationale for testing of JAK inhibitor–based therapies in the clinic.24,44,69,70,93,94 However, preclinical activity of ruxolitinib in PDX models has been somewhat variable, depending on underlying genetic alterations (eg, CRLF2-R vs JAK2 fusion).24 Rigorous clinical testing of JAK inhibition in patients with these specific alterations and elucidation of biomarkers of response and resistance may provide additional insight into these differential responses. Additional therapies of potential relevance for CRLF2-R ALL currently in preclinical testing include USP9X inhibitors, the histone deacetylase inhibitor givinostat, anti-TSLPR/CRLF2 antibodies, and TSLPR-directed chimeric antigen receptor T-cell immunotherapy.67,95-97
The COG AALL1521 phase 2 trial (NCT02723994) is currently investigating the safety and efficacy of combining ruxolitinib with postinduction chemotherapy in children, adolescents, and young adults with newly diagnosed, high-risk JAK pathway–mutant Ph-like ALL (Figure 5). High-risk Ph-like B-ALL patients with CRLF2/JAK pathway lesions are eligible to enter AALL1521 for postinduction therapy combining ruxolitinib with a standard chemotherapy backbone. Patients are stratified by underlying genetic alterations (CRLF2-R, JAK mutations, JAK2 fusions, and EPOR-R) and by end-induction MRD status to determine the potential differential efficacy of combination therapy in each subset. The primary end point of this nonrandomized trial is 3-year event-free survival vs that of historic control patients treated with chemotherapy alone.
In addition to COG AALL1521, a subarm of SJCRH Total XVII is adding ruxolitinib to chemotherapy in children with JAK-mutant Ph-like ALL. The same MDACC trial testing dasatinib in patients with Ph-like ALL and ABL-class fusions is also evaluating ruxolitinib in combination with multiagent chemotherapy in older children and adults with relapsed/refractory JAK-mutant ALL, using complete response rate after 6 weeks of reinduction therapy as the primary end point. Patients with JAK pathway–mutant Ph-like ALL are initially treated with up to 3 weeks of ruxolitinib monotherapy with multiagent chemotherapy added for patients with an incomplete response.
Opportunities for treatment of patients with Ras/MAPK pathway alterations
Up to 6% of patients with Ph-like ALL harbor mutations in the Ras/MAPK signaling pathway (Figure 3) as their sole abnormality.34 Although Ras itself has been impossible to inhibit directly, downstream molecules, such as MEK, are targetable with a class of Food and Drug Administration–approved inhibitors (eg, trametinib, selumetinib, and cobimetinib). Trametinib has been approved for use in BRAF mutant melanoma, and studies are ongoing to define the maximum tolerated dose in children (NCT02124772) with relapsed/refractory solid tumors. The COG ADVL1521 phase 2 trial will test the efficacy of trametinib in juvenile myelomonocytic leukemia, a rare pediatric myeloproliferative neoplasm driven by lesions in the Ras/MAPK pathway.
Future perspectives
Multiple studies have shown that Ph-like ALL is associated with poor survival in both children and adults and have identified a variety of underlying genomic aberrations, many of which can be targeted by commercially available small molecule inhibitors. Anecdotal reports have described the clinical activity of these agents, and single-arm trials have now been initiated that test the addition of targeted agents, including dasatinib and ruxolitinib, to standard chemotherapy backbones. Key questions for the future include how to identify Ph-like ALL patients with targetable genomic lesions in real-time, whether this approach will improve survival, whether there are clinical factors or prognostic biomarkers that can identify patients that will or will not benefit from TKI therapy, the role of allogeneic HSCT in this ALL subset, and whether combinations of targeted therapies will be more effective than a single drug.
Acknowledgments
S.K.T. is supported by National Institutes of Health, National Cancer Institute grant K08CA184418. M.L.L. is the Benioff Chair of Children’s Health at the University of California, San Francisco. S.P.H. is the Jeffrey E. Perelman Distinguished Chair in Pediatrics at The Children's Hospital of Philadelphia.
Authorship
Contribution: S.K.T., M.L.L., and S.P.H. prepared, edited, and approved the manuscript.
Conflict-of-interest disclosure: S.K.T. receives research funding from Gilead Sciences and Incyte Corporation. M.L.L. receives research funding from Incyte Corporation. S.P.H. has received honoraria from Amgen, Jazz Pharmaceuticals, and Erytech and consulting fees from Novartis.
Correspondence: Stephen P. Hunger, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Colket Translational Research Building, Room 3060, 3501 Civic Center Blvd, Philadelphia, PA 19104; e-mail: hungers@email.chop.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal