Key Points
Adult-mBLs have distinct and more frequent DNA copy number abnormalities compared with pediatric-mBL.
Comprehensive genomic analysis revealed that the BCR signaling pathway is a potential therapeutic target in adult-mBL.
Abstract
The adult high-grade B-cell lymphomas sharing molecular features with Burkitt lymphoma (BL) are highly aggressive lymphomas with poor clinical outcome. High-resolution structural and functional genomic analysis of adult Burkitt lymphoma (BL) and high-grade B-cell lymphoma with BL gene signature (adult-molecularly defined BL [mBL]) revealed the MYC-ARF-p53 axis as the primary deregulated pathway. Adult-mBL had either unique or more frequent genomic aberrations (del13q14, del17p, gain8q24, and gain18q21) compared with pediatric-mBL, but shared commonly mutated genes. Mutations in genes promoting the tonic B-cell receptor (BCR)→PI3K pathway (TCF3 and ID3) did not differ by age, whereas effectors of chronic BCR→NF-κB signaling were associated with adult-mBL. A subset of adult-mBL had BCL2 translocation and mutation and elevated BCL2 mRNA and protein expression, but had a mutation profile similar to mBL. These double-hit lymphomas may have arisen from a tumor precursor that acquired both BCL2 and MYC translocations and/or KMT2D (MLL2) mutation. Gain/amplification of MIR17HG and its paralogue loci was observed in 50% of adult-mBL. In vitro studies suggested miR-17∼92’s role in constitutive activation of BCR signaling and sensitivity to ibrutinib. Overall integrative analysis identified an interrelated gene network affected by copy number and mutation, leading to disruption of the p53 pathway and the BCR→PI3K or NF-κB activation, which can be further exploited in vivo by small-molecule inhibitors for effective therapy in adult-mBL.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 1871.
Disclosures
Associate Editor Catherine M. Bollard served as an advisor or consultant for Cellectis. CME questions author Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Alnylam, Biogen, and Pfizer Inc. The authors declare no competing financial interests.
Learning objectives
Upon completion of this activity, participants will be able to:
Compare the copy number abnormalities (CNAs) and mutation profile of adult molecularly defined Burkitt lymphoma (mBL) vs pediatric mBL.
Distinguish recurrent mutations and CNAs in genes regulating phosphoinositide 3-kinase and B-cell receptor signaling in adult mBL.
Determine therapeutic and clinical implications of findings from this genomic analysis of adult mBL.
Release date: October 19, 2017; Expiration date: October 19, 2018
Introduction
Burkitt lymphoma (BL), the most common B-cell non-Hodgkin lymphoma (B-NHL) in childhood, accounts for 30% to 50% of NHLs in this age group. Although representing 1% to 5% of NHLs in adults,1 the absolute number of adult-BL exceeds that of pediatric-BL.2 The World Health Organization recognizes 3 variants: sporadic (sBL), endemic (eBL), and BL associated with HIV infection.3 Effective treatment of BL involves high-dose intensive chemotherapeutic regimens with a more than 80% probability of 5-year event-free survival in pediatric patients,4 but similar improvement in clinical outcome has not yet been achieved in patients with adult-BL, particularly in those older than 40 years.5
Gene expression profiling (GEP),6,7 immunoglobulin (Ig) mutation, and immunophenotypic analysis indicate a germinal center (GC) B-cell origin. The genetic hallmark is MYC-IGH translocation [t(8;14)(q24;q32)], which occurs in 80% of cases. MYC-IGL translocations [t(8;22) or t(2;8)] occur in 10% of BL.8 Approximately 10% of adult and 30% of pediatric cases of diffuse large B-cell lymphoma (DLBCL) also have MYC translocation, which may lead to misdiagnosis.9 This diagnostic distinction is critical because aggressive chemotherapeutic regimens are curative in BL, but standard regimens for DLBCL are inferior.
Genomic studies have uncovered novel disease-related abnormalities and refined pathogenic mechanisms that have assisted in more accurate BL diagnosis.6,7 However, most studies have focused on pediatric-BL,10-12 with few comprehensive genomic studies in adult-BL. Array comparative genomic hybridization techniques revealed recurrent 1q21.1-q31.3 and 13q31 gains and 17p deletion in BL, but other aberrations were inconsistent.11,12 Therefore, we conducted an integrative analysis of the genomic copy number abnormalities (CNA), global GEP, and mutation analysis on adult cases (age >21 years) who were pathologically diagnosed as BL, Burkitt-like lymphoma, or DLBCL, but defined as BL by GEP, using a previously derived molecular signature.6,7 These cases were designated molecularly defined BL (mBL). We compared the CNA and mutation profile of these cases with pediatric-mBL and other GC-derived B-NHL, including follicular lymphoma (FL), transformed FL (tFL), and de novo GCB-DLBCL. We identified recurrent mutations and CNA in genes regulating phosphoinositide 3-kinase (PI3K) and B-cell receptor (BCR) signaling in adult-mBL, which can be targeted by ibrutinib. This study illustrates that comprehensive genomic analysis can identify oncogenic pathways and reveal promising targets for novel therapeutic intervention in adult-mBL.
Methods
Detailed materials and methods are available in the supplemental Materials and methods, available on the Blood Web site.
Patient samples and B-cell lines
Select clinical characteristics are provided in Tables 1 and 2 and supplemental Table 1A. Genomic data from published series of mBL (12 adults, 18 pediatric), FL, tFL,13 and GCB-DLBCL14,15 were included for comparative analysis. The genetic characteristics of BL cell lines are tabulated in supplemental Table 1B, and cell culture details are in the supplemental Materials and methods.
Clinical characteristics: molecular BL cases
| . | . | . | Path diagnosis . | |||
|---|---|---|---|---|---|---|
| Adult . | Pediatric . | |||||
| . | Adult . | Pediatric . | BL . | DLBCL . | BL . | DLBCL . |
| Current study | ||||||
| Total | 22 | 39 | 18 | 4 | 36 | 3 |
| Male | 14 | 32 | 12 | 2 | 30 | 2 |
| Female | 8 | 7 | 6 | 2 | 6 | 1 |
| Average age (range) | 56.6 (27-82) | 9.2 (3-20) | ||||
| GSE21597 | ||||||
| Total | 9 | 13 | ||||
| Male | 5 | 12 | ||||
| Female | 4 | 1 | ||||
| Average age, y (range) | 41 (24-63) | 7.3 (2-13) | ||||
| Total | 28 | 45 | ||||
| . | . | . | Path diagnosis . | |||
|---|---|---|---|---|---|---|
| Adult . | Pediatric . | |||||
| . | Adult . | Pediatric . | BL . | DLBCL . | BL . | DLBCL . |
| Current study | ||||||
| Total | 22 | 39 | 18 | 4 | 36 | 3 |
| Male | 14 | 32 | 12 | 2 | 30 | 2 |
| Female | 8 | 7 | 6 | 2 | 6 | 1 |
| Average age (range) | 56.6 (27-82) | 9.2 (3-20) | ||||
| GSE21597 | ||||||
| Total | 9 | 13 | ||||
| Male | 5 | 12 | ||||
| Female | 4 | 1 | ||||
| Average age, y (range) | 41 (24-63) | 7.3 (2-13) | ||||
| Total | 28 | 45 | ||||
Clinical characteristics: nonmolecular BL cases (BL by pathology diagnosis), n = 17
| . | . | . | Path diagnosis . | |||||
|---|---|---|---|---|---|---|---|---|
| DLBCL subtype . | ||||||||
| Adult . | Pediatric . | |||||||
| ABC . | GCB . | UC . | ABC . | GCB . | UC . | |||
| Current study | ||||||||
| Total | 7 | 10 | 5 | 1 | 1 | 1 | 7 | 2 |
| Male | 5 | 7 | 3 | 1 | 5 | 1 | ||
| Female | 2 | 3 | 2 | 1 | 1 | 2 | 1 | |
| Average age, y (range) | 76.29 (62-85) | 6.4 (2-11) | ||||||
| . | . | . | Path diagnosis . | |||||
|---|---|---|---|---|---|---|---|---|
| DLBCL subtype . | ||||||||
| Adult . | Pediatric . | |||||||
| ABC . | GCB . | UC . | ABC . | GCB . | UC . | |||
| Current study | ||||||||
| Total | 7 | 10 | 5 | 1 | 1 | 1 | 7 | 2 |
| Male | 5 | 7 | 3 | 1 | 5 | 1 | ||
| Female | 2 | 3 | 2 | 1 | 1 | 2 | 1 | |
| Average age, y (range) | 76.29 (62-85) | 6.4 (2-11) | ||||||
Structural and functional genomic analysis
Comparative genomic hybridization and copy number analysis.
Target gene section and whole-exome capture and data analysis.
A panel (n = 380) targeting genes mutated (≥5%) in NHLs was designed. Libraries were prepared from genomic DNA, and exomes were captured with the Kapa Biosystems exome enrichment kit per manufacturer's protocol (Kapa Biosystems, Inc). Data analysis was performed as previously described16 and detailed in the supplemental Materials and methods.
Gene and miRNA expression analysis.
The GEP data from a previous study6 were used to classify samples as BL or DLBCL, using a Bayesian classifier.6 Differential gene expression and association with CNA status (gain, loss, normal) were analyzed by Student t test, using approaches described in earlier studies.9 miRNA expression was generated previously,17 using the TaqMan human microRNA array.
MYC and BCL2 protein expression and translocation status
Tissue microarrays were stained with MYC and BCL2 antibodies, as described previously.18,19 More than 50% staining was the positive threshold. Supplemental Table 7 summarizes cases with MYC or the BCL2 translocation identified by sequencing, fluorescence in situ hybridization, and validation by long-range polymerase chain reaction.20
Survival outcome analysis
Overall survival was estimated using the Kaplan-Meier method, and differences were assessed using the log-rank test.
Functional validation using in vitro assays in BL cell lines
Quantitation of MYC and miRNA17∼92.
Expression of MYC and miR17∼92 was estimated by quantitative reverse transcription polymerase chain reaction (primers in supplemental Table 8), and microRNA 17∼92 (miR17∼92) cluster members were evaluated by TaqMan small RNA assays.
MYC and miRNA17∼92 knockout/knockdown in cell lines.
MYC and MIR17HG knockout was performed using CRISPR-mediated gene editing (single-guide RNA sequences listed in supplemental Table 8). Functional loss of miR17∼92 was performed using a previously described miRNA sponge.21 Sponge efficiency was estimated by expression of miRNA17∼92 targets (BIM and PTEN).
Evaluation of BCR signaling and in vitro ibrutinib treatment.
BCR activation status was estimated by the phosphorylation status of upstream regulators (BLNK, BTK, and SYK) and a downstream effector (ERK) after IgM treatment and BCR inhibition was performed using ibrutinib, as detailed in the supplemental Data (supplemental Figure 11).
Cell proliferation.
Cell proliferation was estimated using Presto Blue/CyQUANT (Thermo Fisher, Inc). Apoptosis rate was calculated using FACSCalibur (BD Biosciences) after Annexin-V staining (BioLegend, Inc). Cell cycle was analyzed with a BD LSR II flow cytometer after propidium iodide staining.
Results
Patient characteristics and molecular classification of BL by GEP
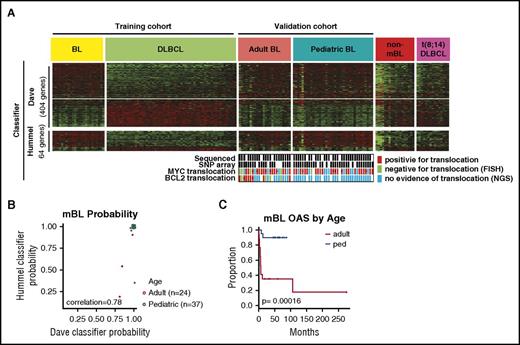
Clinical characteristics of the 61 mBL cases by GEP are summarized in Tables 1 and 2. Figure 1A-B depicts expression of the previously defined molecular BL signature6 and validation of the classification using a different molecular-BL classifier.7 Pediatric-mBL and adult-mBL cases were indistinguishable by the classifier and distinct from cases diagnosed as BL by pathology, but molecularly classified as DLBCL or unclassifiable (non-mBL) and t(8;14)-positive DLBCL22 (Figure 1A). Examination of the molecular classifier in other BL series (GSE6905323 and GSE44757 ) confirmed that adult-mBL and pediatric-mBL showed concordant gene signatures (supplemental Figure 1A-D). Patients with adult-mBL, however, had significantly worse overall survival5 (Figure 1C), as also noted in a previous study23 (supplemental Figure 1D). Adult and pediatric patients with mBL had a similar proportion of t(8;14)-positive cases and similar morphological features, but on genomic analysis, several genetic characteristics differed significantly (supplemental Figure 1E; supplemental Table 1A).
Adult and pediatric-mBL cases are indistinguishable by the Burkitt classifier but have distinct CNAs. (A) Heat map showing expression of genes in the Dave et al6 and Hummel et al7 Burkitt classifiers for a BL and DLBCL training cohort and a validation cohort consisting of adult-BL, pediatric-BL, nonmolecularly classified Burkitt, and t(8;14)-positive DLBCL tumors. MYC and BCL2 translocation status of the 61 cases in the study are noted, and cases in which the translocation status comes solely from NGS data are marked (·). (B) Comparison of the mBL probability score predicted by the Dave et al6 and Hummel et al7 BL classifiers. Three cases classified as Burkitt by the Dave classifier would be classified as intermediate by the Hummel classifier. (C) Kaplan-Meier curves comparing overall survival of adult and pediatric molecularly classified Burkitt cases. The adult-BL series shows dismal clinical outcome, which may be attributed to suboptimal treatment, diagnostic challenges (before the GEP era), double-hit cases with mBL features, and advanced age (median, 55 years).
Adult and pediatric-mBL cases are indistinguishable by the Burkitt classifier but have distinct CNAs. (A) Heat map showing expression of genes in the Dave et al6 and Hummel et al7 Burkitt classifiers for a BL and DLBCL training cohort and a validation cohort consisting of adult-BL, pediatric-BL, nonmolecularly classified Burkitt, and t(8;14)-positive DLBCL tumors. MYC and BCL2 translocation status of the 61 cases in the study are noted, and cases in which the translocation status comes solely from NGS data are marked (·). (B) Comparison of the mBL probability score predicted by the Dave et al6 and Hummel et al7 BL classifiers. Three cases classified as Burkitt by the Dave classifier would be classified as intermediate by the Hummel classifier. (C) Kaplan-Meier curves comparing overall survival of adult and pediatric molecularly classified Burkitt cases. The adult-BL series shows dismal clinical outcome, which may be attributed to suboptimal treatment, diagnostic challenges (before the GEP era), double-hit cases with mBL features, and advanced age (median, 55 years).
Structural genomic (CNA and mutation) analysis of mBL and other GC-derived B-NHLs
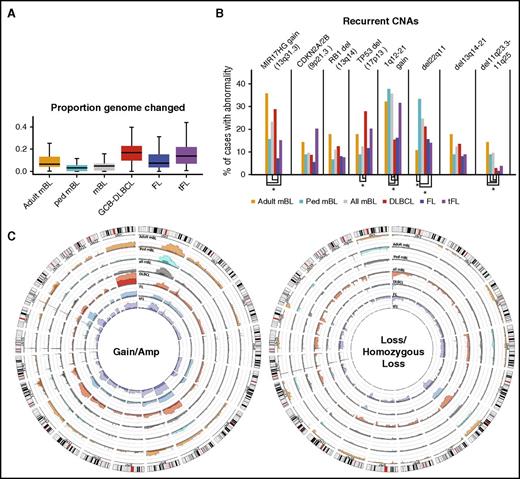
A total of 73 cases were included for CNA analysis (51+22 mBL cases from an earlier study10 ). The 2 cohorts showed similar genomic profiles (supplemental Figure 2A) and were combined to identify recurrent CNA. Comparison of the mBL cohort with other GC-derived B-NHL (ie, FL, tFL, and de novo GCB-DLBCL)13,14 showed that a smaller percentage of the mBL genome (5.7%) was aberrant compared with other GC-derived B-NHLs (FL, 11%; tFL, 17%; GCB-DLBCL, 17%; Figure 2A), consistent with cytogenetic studies.8 Most chromosomes showed some degree of aberration in mBL, with a few loci unique (del11q23.3-11q25) or more frequently (del13q14-21, del22q11) or more focally (13q31.3 gain) altered in mBL compared with FL, tFL, and GCB-DLBCL (Figure 2B-C; supplemental Figure 2B-C). Despite the low genomic complexity of the mBL genome, some aberrations were as frequent as 20% to 35% (1q12-21 gain, 13q31.3 gain, cnLOH-17q; Figure 2B-C; supplemental Figure 2B-C,E). Loss of genomic regions encompassing well-characterized tumor suppressors (TS) or cell cycle regulators, including TP53 (17p13.1), RB1 (13q14.2), and CDKN2A/2B (9p21.3), was common in mBL, although not as frequent as in other NHLs. However, del22q11, del11q23.3-11q25, and focal gain of 13q31.3 occurred more frequently in mBL than in other GC-derived B-NHLs.
CNAs in germinal center B-cell-derived lymphomas. (A) Box plot of the proportion of the aberrant genome (change in genome content) for the noted GC-B-cell-derived NHL entities. (B) Bar graph of the frequency or recurrent CNAs in different lymphomas. CNAs with significantly a different (P < .05) frequency of CNAs are denoted (*). (C) Circos plots comparing the frequency of gains and losses found in the noted lymphoma types. Darker shades denote regions of amplifications or homozygous copy loss and axis lines represent 20%.
CNAs in germinal center B-cell-derived lymphomas. (A) Box plot of the proportion of the aberrant genome (change in genome content) for the noted GC-B-cell-derived NHL entities. (B) Bar graph of the frequency or recurrent CNAs in different lymphomas. CNAs with significantly a different (P < .05) frequency of CNAs are denoted (*). (C) Circos plots comparing the frequency of gains and losses found in the noted lymphoma types. Darker shades denote regions of amplifications or homozygous copy loss and axis lines represent 20%.
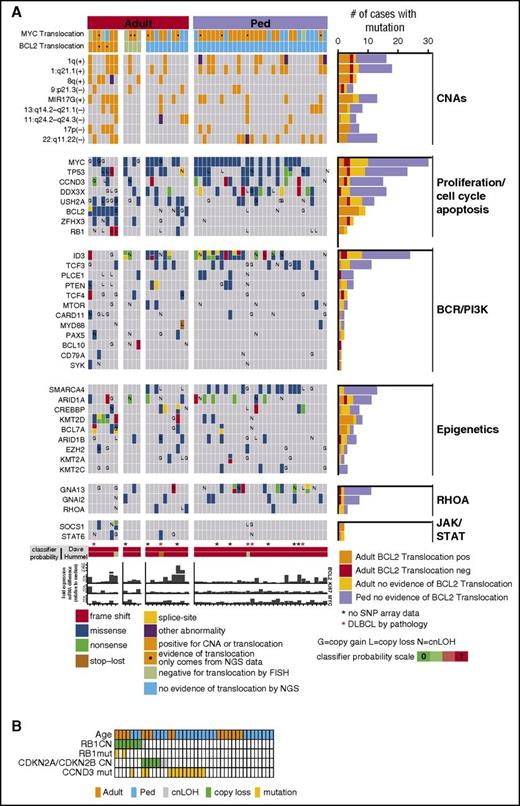
Mutation data were generated for 52 mBLs (Figure 3A; supplemental Tables 2 and 3), and MYC (58%), ID3 (46%), TP53 (44%), DDX3X (31%), CCND3 (29%), SMARCA4 (25%), and TCF3 (21%) were mutated frequently in mBL, more so than in other GC-derived B-NHLs24 (supplemental Figure 2F), and similar to earlier BL studies25-27 (supplemental Figure 2G). Several mutated genes (TCF3, CDH23, CSMD2, BCL7A, and EPPK1) were unique to BL, and genes associated with the chromatin remodeling complex SWI/SNF (SMARCA4, ARID1A, and ARID1B) were frequently mutated in mBL. RYR1, ARID1B, BCL7A, and PLCE1 mutations, present in 10% of mBL, have not been previously reported.25-27
Select CNA or genes found to be recurrently mutated in 52 mBL cases. (A) The block color represents the type of mutation. Blocks with 2 colors indicate that more than 1 type of mutation was observed. Genes also affected by copy number abnormalities are noted (G, copy gain; L, copy loss; N, copy neutral loss of heterozygosity). MYC and BCL2 translocation status is shown and MYC, BCL2, and KI67 mRNA expression relative to the median is depicted. (B) Copy number and/or mutation status of RB1, CDKN2A/CDKN2B, and CCND3 for individual mBL tumors.
Select CNA or genes found to be recurrently mutated in 52 mBL cases. (A) The block color represents the type of mutation. Blocks with 2 colors indicate that more than 1 type of mutation was observed. Genes also affected by copy number abnormalities are noted (G, copy gain; L, copy loss; N, copy neutral loss of heterozygosity). MYC and BCL2 translocation status is shown and MYC, BCL2, and KI67 mRNA expression relative to the median is depicted. (B) Copy number and/or mutation status of RB1, CDKN2A/CDKN2B, and CCND3 for individual mBL tumors.
MYC mutations were never observed in the DNA binding domain (supplemental Figure 3A), but were confined to the activation domain within the MYC box I and PEST domains and at a hotspot affecting a proline residue (MYCP72/57), whose mutation results in inhibition of ubiquitin-mediated degradation.28 No association with MYC mRNA or CN status was observed with mutation status. TP53 mutations occurred only in the DNA binding domain (supplemental Figure 3B) and included hotspot mutations affecting R248 (n = 4) and R273 (n = 2).29 Several TP53 mutations were accompanied by copy neutral loss of heterozygosity (cnLOH) or copy loss. Cases with copy loss or cnLOH have significantly lower expression of TP53 than those without mutation or CNAs (∼2-fold change P = .04; supplemental Figure 4A). ID3 mutations occurred in the helix-loop-helix domain and were accompanied by LOH of ID3 (supplemental Figure 3C; Figure 3A).25-27 Another ID3 mutation affecting a splice-donor site, reported to result in deletion of amino acids 82-100,27 occurred in several cases. TCF3 mutations were located in the helix-loop-helix domain (supplemental Figure 3E) and likely interfere with regulation by ID3.27 Correlation of GEP and mutation status (supplemental Figure 4B) indicated that cases with ID3 mutation expressed higher levels of ID3 mRNA, in agreement with a previous study.25 Consistent with previous studies,26,27 most CCND3 mutations affected the carboxy terminus with 2 hotspots (I290 and T271; supplemental Figure 3D). Several other genes showed differential mRNA expression based on mutation status (supplemental Figure 4B).
Structural genomic (CNA and mutation) analysis in adult and pediatric-mBL
A higher proportion of the BL genome was aberrant in adult compared with pediatric cases (Figure 2A; supplemental Table 4). Figure 2C and supplemental Figure 2D illustrate that adult-mBLs have a higher frequency of abnormalities at any genomic locus except 22q11.22.
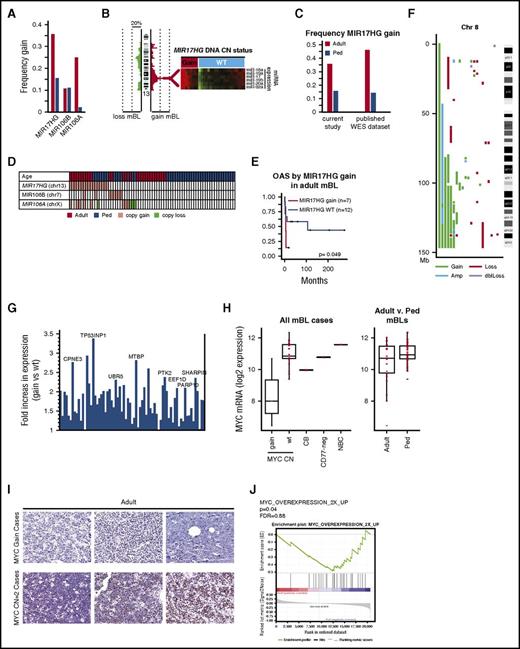
13q31.3 gain, affecting MIR17HG(miR17∼92 cluster) occurred more than twice as frequently in adult as in pediatric cases (36% vs 16%; Figure 4A; Fisher’s exact test P = .08), and was associated with increased expression of miR17∼92 members, as measured by quantitative reverse transcription polymerase chain reaction (Figure 4B). The frequent MIR17HG gain in adult BL was validated in a separate BL cohort (n = 27) profiled using whole exome sequencing (WES)25 (Figure 4C). miR17∼92 paralogues (miR106b∼25 at 7q22.1, and miR106a∼363 at Xp26.2), which share the same seed sequence, were affected by copy gains, with miR106a∼363 more frequent in adult-mBLs (Figure 4A). Gain/amplification of miR17∼92 or paralogues occurred in 50% (14/28) of adult-BL compared with 27% (12/45) of pediatric-BL (Figure 4D; Fisher's exact test P = .05). Although miRNA expression was not increased in cases with gain of the paralogues, expression of miR17∼92 and its paralogues was higher in mBL than in DLBCL (supplemental Figure 5A-B). Adult-mBL patients with MIR17HG gain had worse overall survival (Figure 4E), and gene set enrichment analysis identified enrichment of gene signatures associated with immune suppression or hypoxia (eg, transforming growth factor β and hypoxia inducible factor 1-α target signature) and chemotherapy resistance in adult cases with MIR17HG gain (supplemental Figure 5C). In contrast, MYC and E2F target signatures and proliferation signatures were enriched in MIR17HG WT cases (supplemental Figure 5C).
MIR17HG and Chr8q are gained in mBL. (A) Frequency of DNA CN gains found in adult (red-orange) or pediatric (blue) mBL for MIR17HG and paralogues MIR106B and MIR106A. (B) Heat maps of expression of the MIR17∼92 cluster members measured by TaqMan Array Human MicoRNA A Card (ABI) in tumors with and without MIR17HG DNA copy number gain. (C) Validation of the frequency of MIR17HG DNA CN gain in adult-BL using a published BL genomic/WES dataset. CN status was assessed using Varscan2 CNV followed by segmentation by the Circular binary segmentation algorithm. For WES data, regions of gain were cut off at a threshold of 0.2 log2 ratio. (D) Copy number status of MIR17HG and paralogues for individual tumors (gain = red, loss = green). (E) Kaplan-Meier curves comparing overall survival adult-mBL cases by MIR17HG copy number status. Several MIR17HG gain/amplified cases also had double MYC/ BCL2 translocation/mutation, but showed BL mutation and GEP profile. (F) Plot of CNAs along chromosome 8 for mBL cases with abnormalities. (G) Bar graph of the fold increase in expression of genes along chromosome 8 that are significantly upregulated (P < .05, 1-sided Student t test) in cases with CN gain. Genes of interest are noted. (H) Boxplot of MYC mRNA expression by MYC DNA copy number status and by age. Red points represent expression values of individual cases. Expression levels in normal centroblasts (CB), CD77-negative cells, and naive B cells (NBC) are noted for comparison. (I) MYC protein expression in 3 cases with MYC gain and 3 cases with a MYC DNA copy number=2. (J) Signatures upregulated on MYC overexpression were enriched in pediatric-mBLs compared with adult-mBLs.
MIR17HG and Chr8q are gained in mBL. (A) Frequency of DNA CN gains found in adult (red-orange) or pediatric (blue) mBL for MIR17HG and paralogues MIR106B and MIR106A. (B) Heat maps of expression of the MIR17∼92 cluster members measured by TaqMan Array Human MicoRNA A Card (ABI) in tumors with and without MIR17HG DNA copy number gain. (C) Validation of the frequency of MIR17HG DNA CN gain in adult-BL using a published BL genomic/WES dataset. CN status was assessed using Varscan2 CNV followed by segmentation by the Circular binary segmentation algorithm. For WES data, regions of gain were cut off at a threshold of 0.2 log2 ratio. (D) Copy number status of MIR17HG and paralogues for individual tumors (gain = red, loss = green). (E) Kaplan-Meier curves comparing overall survival adult-mBL cases by MIR17HG copy number status. Several MIR17HG gain/amplified cases also had double MYC/ BCL2 translocation/mutation, but showed BL mutation and GEP profile. (F) Plot of CNAs along chromosome 8 for mBL cases with abnormalities. (G) Bar graph of the fold increase in expression of genes along chromosome 8 that are significantly upregulated (P < .05, 1-sided Student t test) in cases with CN gain. Genes of interest are noted. (H) Boxplot of MYC mRNA expression by MYC DNA copy number status and by age. Red points represent expression values of individual cases. Expression levels in normal centroblasts (CB), CD77-negative cells, and naive B cells (NBC) are noted for comparison. (I) MYC protein expression in 3 cases with MYC gain and 3 cases with a MYC DNA copy number=2. (J) Signatures upregulated on MYC overexpression were enriched in pediatric-mBLs compared with adult-mBLs.
8q22-q24 gain was only observed in adult-mBL (30%) (Figure 4F). Several genes showed increased expression with CN gain, including MTBP, a likely MYC transcriptional target, MYC-interacting protein, and regulator of proliferation30 (Figure 4G), and UBR5, an E3 ubiquitin ligase mutated in 2 adult-mBL cases and known to be affected by CNA/mutation in tumors.27 Unexpectedly, cases with MYC gain (8q) had low MYC mRNA expression compared with wild-type cases (>2-fold change, P < .001; Figure 4H) and low protein expression by immunohistochemistry (0/4 MYC positive in 8q24 gain vs 6/10 MYC positive in 8q24-WT; P = .085; Figure 4I). MYC mRNA expression was higher in pediatric compared with adult-mBL (mRNA, P = .014; Figure 4H), more pediatric cases were positive for MYC protein expression (7/10 in pediatric vs 6/15 in adult), and enrichment of MYC target gene expression was observed in pediatric-mBL compared with adult-mBL (Figure 4J).
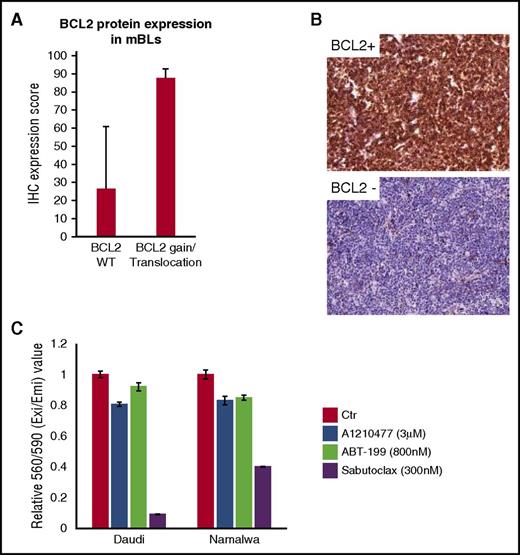
18q gain was more frequent in adult-mBL than pediatric-mBL (Figure 2C; supplemental Figure 2D; 18% vs 9%), and genes on 18q with a concomitant increase in mRNA expression with CNA included BCL2, MALT1, TCF4, SOCS6, and PIAS2 (supplemental Figure 6F). BCL2 mRNA was ∼4 fold higher in cases with 18q gain, and BCL2 gain/translocation was associated with BCL2 protein expression (supplemental Figures 5A-B and 6F). Another BCL2 family member, MCL1 at chr1q21 crucial for MYC-driven lymphoma maintenance,31 showed similar frequency of CN gain in adult and pediatric-mBL (∼35%), and did co-occur with BCL2 aberrations. BCL2 and MCL1 coexpression occurred in BL cell lines (supplemental Figure 10C), and they were more sensitive to the pan-BCL2/MCL-1/BCL-XL/BCL2A1 inhibitor Sabutoclax than to selective BCL2 (ABT-199) or MCL1 (A1210477) inhibitors (Figure 5C).
18q21 gain or BCL2 translocation is associated with BCL2 protein expression. (A) BCL2 IHC was performed and scored on adult-mBL cases with no evidence of BCL2 gain/translocation based on clinical data, NGS, and SNP array (n = 7) and cases with BCL2 gain/translocation (n = 6). Bars represent the mean expression score and error bars represent 1 standard deviation. (B) Example of BCL2-positive and BCL2-negative staining cases (original magnification, ×20). (C) BL cell lines are sensitive to a pan-BCL2 inhibitor compared with single-agent inhibitors. Bars represent the mean of 3 replicates from 1 representative experiment of at least 3 separate experiments, and error bars represent 1 standard deviation.
18q21 gain or BCL2 translocation is associated with BCL2 protein expression. (A) BCL2 IHC was performed and scored on adult-mBL cases with no evidence of BCL2 gain/translocation based on clinical data, NGS, and SNP array (n = 7) and cases with BCL2 gain/translocation (n = 6). Bars represent the mean expression score and error bars represent 1 standard deviation. (B) Example of BCL2-positive and BCL2-negative staining cases (original magnification, ×20). (C) BL cell lines are sensitive to a pan-BCL2 inhibitor compared with single-agent inhibitors. Bars represent the mean of 3 replicates from 1 representative experiment of at least 3 separate experiments, and error bars represent 1 standard deviation.
Loss of genomic loci (6q21-25, 9p21.3, 13q14-q21.2, and 17p13.1) encompassing well-characterized TS genes was frequent (∼10%-20%) in adult-mBL. Del17p13.1 affecting TP53 was twice as frequent in adult-mBLs as in pediatric-mBLs (18% vs 9%); however, TP53 cnLOH and/or mutation occurred at equal frequency (11% cnLOH and ∼45% mutation). Focal del9p21.3 encompassing CDKN2A/CDKN2B (14% adult, 9% pediatric) and del13q14.2 affecting RB1 (18% adult and 7% pediatric) were found. RB1 mutations were only identified in adult-mBL (∼15%, 3/21), and cases with RB1 copy loss and/or mutation had an average 3-fold or higher decrease in mRNA expression compared with WT cases. Although CDKN2A/CDKN2B loss and RB1 copy loss/mutation did not co-occur, the sample number was insufficient to determine the significance of mutual exclusivity (Figure 3B).
1q21-q25.3 gain/amplification, proximal to the centromere, was present with similar frequency (∼35% to 40%) in adult and pediatric-mBL, but the frequency of the distal loci (1q25.3-1q42.13 gain) was higher in adults (30% to 40% vs 10% to 25%, Figure 2C; supplemental Figure 2D), and included genes whose encoded proteins negatively regulate p53 stability (MDM4), ubiquitinate MYC (FBXO28), or are primarily associated with proliferation/survival (RGS16, BTG2, EIF2D CENPF, ASPM) and gene expression levels correlated with CN status (P < .05 and >1.5-fold change; supplemental Figure 6A). In addition, 1q25 gain encompassing MDM4 (36% adult, 18% pediatric) and 12q15 gain containing MDM2 (14% adult, 4% pediatric), were also noted in mBL; gene expression showed that CN gain led to higher mRNA expression (>2-fold, P < .01). Additional genes within recurrently aberrant loci had associated gene expression changes and may contribute to lymphomagenesis (supplemental Figure 6A-F).
Mutational landscape in adult-mBL and pediatric-mBL
The frequencies of commonly mutated genes (eg, MYC, ID3, TP53, CCND3, DDX3X, ARID1A, and TCF3) were not significantly different in adult-mBL and pediatric-mBL (Figure 3A; supplemental Tables 2 and 3). However, BCL2 (43% vs 0; P < .001), ZFHX3 (24%; P < .01), SPTBN5 (20%; P = .02), RB1 (14%; P = .06), BTG1 (14%; P = .06), TCF4 (14%; P = .06), and TNFRSF14 (14%; P = .06) were exclusively mutated in adult-mBL. In contrast, mutations in CDH23 (29% vs 5%; P = .04), SMARCA4 (35% vs 19%; P = .05), RYR3 (13% vs 0; P = .14), and RYR1 (29% vs 10%; P = .16) were more frequent in pediatric-mBL. Of note, SMARCA4 (25% all mBL) and KMT2D (15% all mBLs) mutations were mutually exclusive (P = .04, CoMEt33 ). Two Gα subunits of G-protein-coupled receptors, GNA13 and GNAI2, were mutated more frequently in pediatric-mBL (29% vs 9.5% and 16% vs 9.5%, respectively), and mutation only co-occurred in 1 sample (Figure 3A).
Analysis of coding and noncoding mutations in genes known to be targeted by activation induced cytidine deaminase34 showed more mutations/case in adult-mBLs compared with pediatric-mBLs (29 vs 15). Cases with known MYC or BCL2 translocations had more noncoding MYC and BCL2 mutation, respectively (supplemental Figure 7A). Cases with 10 or more mutations in MYC were frequent in pediatric-mBL (77% vs 33%). In contrast, more than 10 mutations in BCL2 or BCL6 occurred more frequently in adult-mBL (BCL2: 33% vs 0%; BCL6: 24% vs 3%; supplemental Figure 7B). No coding mutation was observed in BCL6 in adult-BL.
Mutations and CNA define major oncogenic pathways in adult-mBL
Mutations in genes regulating the PI3K-AKT pathway (TCF3, ID3, PTEN, and PLCE1) did not show a significant difference between adult and pediatric cases. However, mutations in BCR signaling effectors that induce chronic active NF-κB signaling (CD79A, SYK, MYD88, BCL10, and CARD11) were more frequent in adult-mBL (any mutation: 19% vs 7%). Several genes involved in NF-κB pathway activation, including MALT1 at 18q, BCL11A and REL at 2p16, and TRAF5 and TRAF3IP3s at 1q32, were aberrantly gained in adult-mBL, with concomitant upregulated mRNA expression.
Genes associated with mature B-cell development/differentiation (IRF8 and PAX5) were mutated only in adult-BL (either mutation: 19% vs 0), in contrast to genes associated with GC differentiation (BCL6 [coding], NOTCH1, NOTCH2), which were found in pediatric-mBL (16% of pediatric cases with any mutation vs none). Mutations that regulate the S1P/S1PR pathway (GNA13, GNAI2) were primarily observed in pediatric-BL (Figure 3A). Many GNAI2 mutations were missense mutations; a recent study suggested that mutations in GNAI2 may be gain-of-function.35 GNA13 couples to S1PR2 and results in inhibition of PI3K/AKT signaling, whereas GNAI2 couples to S1PR1 and promotes PI3K/AKT signaling.
We observed differences in gene signatures and signaling pathways between adult and pediatric-mBL (supplemental Figure 8A).36 Further analysis using gene set enrichment analysis37 identified enrichment of STAT3 targets and ubiquitin-mediated proteolysis signatures in adults, whereas the S1PR pathway and genes induced on MYC overexpression were enriched in pediatric-mBL (supplemental Figure 8B). Many enriched gene signatures or pathways in adults are likely a result of differences in the frequency of MIR17HG gain, or mutations unique in these 2 age groups. For example, JAK-STAT pathway mutations occurred only in adults, whereas GNA13 and GNA12 mutations were primarily in pediatric-mBL. Enrichment of MYC-regulated genes may be a result of MYC expression differences between the 2 groups (Figure 4H,J). Differentially expressed genes in adult-mBL (vs pediatric-BL) showed similarity to adult NHL, as well as BL cell lines, even though the majority of these cell lines are derived from pediatric patients (supplemental Figure 8A).
Integrative analysis of genomic abnormalities with gene expression
Genes with significant expression changes with CNA status (reduced expression with loss or increased expression with gain) were compiled, and DAVID (https://david.ncifcrf.gov/) showed enrichment of genes involved in cell cycle, the ubiquitin pathway, and TP53 signaling (supplemental Table 5).
Hierarchical clustering of recurrent CNAs and mutations (present in ≥10% of mBL) showed that cases clustered into 2 main groups, dominated by the 2 age subgroups (supplemental Figure 9A). Abnormalities more frequent in the adult cases formed 1 cluster, whereas genetic events frequent in pediatric cases (SMARCA4, GNA13, DDX3X, and OBSCN) formed another cluster with only few cases interspersed, suggesting distinct oncogenic mechanisms in the age-defined subgroups.
We analyzed the co-occurrence of the most frequent genetic alterations, using Pearson correlation, and observed a negative correlation of MIR17HG copy gain with chromosome 17p-cnLOH and SMARC4A mutation (supplemental Figure 9B). TP53 mutation and del17p13.2 were positively correlated (supplemental Figure 9B). TP53 mutations were detected in all cases with copy loss or cnLOH, but several mBL cases only had TP53 mutation (Figure 3A).
Functional losses of MYC and miRNA17∼92 inhibit proliferation
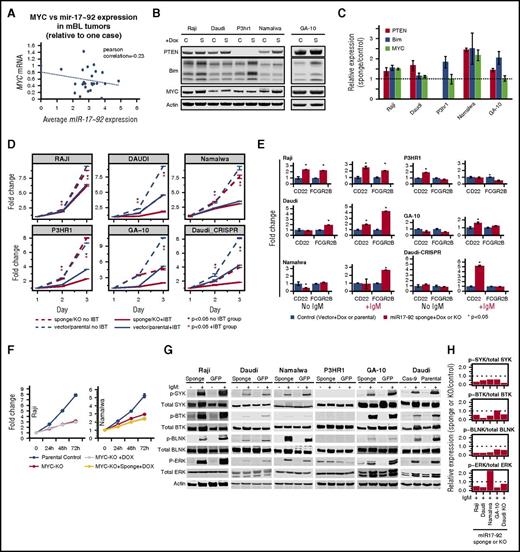
MIR17HG gain, despite being a bona fide MYC target, showed negative correlation (r = −0.23) with MYC expression in tumors (noted in an earlier study12 ; Figure 6A), although a feedback loop fine-tuning MYC expression by miR17∼92 has recently been demonstrated in vitro.38 Significantly higher miRNA expression of miR17∼92 cluster members in BL than in DLBCL (supplemental Figure 5B) or normal GC-B cells17 suggested a distinct role of miR17∼92 in BL pathogenesis. To validate this observation further through in vitro analysis, we examined MIR17HG/miR17∼92 and MYC status (CN, mRNA) in 6 BL cell lines, including 1 from an adult (GA-10).39 Three cell lines (Namalwa, Daudi, Ramos) had MIR17HG gain/amplification and 3 (Namalwa, Raji, P3HR1) had MYC gain/amplification. Despite having normal MYC and MIR17HG CN status, GA-10 showed high mRNA expression of MYC and miR17∼92 cluster members (miR-17, miR-19b, and miR-92a), comparable to those with CN gain (supplemental Figure 10A). Unlike primary tumors, MYC CN status and mRNA expression were positively correlated in cell lines (supplemental Figure 10B). BIM, a miR17∼92 target, had lower expression in cell lines with MIR17HG gain (supplemental Figure 10C).
Evaluation of functional significance of MYC and MIR17HG in BL tumors and cell lines. (A) Comparison of average miR-17-92 expression and MYC expression in mBL tumors. miR17∼92 expression levels were inversely correlated with MYC expression in tumors (r = −0.23). (B) Whole-cell lysates were western blotted for PTEN, BIM, MYC, and β-actin expression in the noted BL cell lines expressing empty vector (C) or a vector containing mIR17-92 sponge (S). (C) BIM, PTEN, and MYC expression was normalized to β-actin, and relative expression (sponge/vector control) was quantified from 2 separate western blot experiments. Error bars represent 1 standard deviation. (D) Proliferation in BL cell lines after miR17∼92 sponge expression or MIR17HG knock out (Daudi-CRISPR) and/or ibrutinib (IBT) treatment. Line graph depicts 1 representative experiment of triplicate experiments, and points are the average of 3 biological replicates. Error bars represent 1 standard deviation. BL cell lines were treated with a specific concentration of IBT based on the 50% infective dose calculation for each line. (E) Comparison of mRNA levels of CD22 and FCGR2B in control (vector or parental) or miR17∼92 sponge or knockout (Daudi-CRISPR) cells with or without IgM stimulation. Fold change was calculated relative to the vector or parental controls. Graph depicts 1 representative experiment of 3 separate experiments. Error bars depict 1 standard deviation of 3 technical replicates. (F) Cell proliferation in BL cell lines after MYC KO and/or miR17∼92 knockdown (sponge). Points represent the mean of 3 biological replicates from 1 representative experiment of at least 3 separate experiments. Error bars represent the standard error of the mean. (G) Western blots of whole-cell lysates from control (GFP or parental) or miR17∼92 sponge-expressing or MIR17HG knock-out (Daudi-Cas-9) BL cell lines after induction of BCR signaling with anti-IgM. (H) Relative expression of phosphorylated/total SYK, BTK, BLNK, and ERK was quantified from the western blot, suggesting decreased BCR signaling in miR17∼92 sponge-expressing or KO BL cell lines.
Evaluation of functional significance of MYC and MIR17HG in BL tumors and cell lines. (A) Comparison of average miR-17-92 expression and MYC expression in mBL tumors. miR17∼92 expression levels were inversely correlated with MYC expression in tumors (r = −0.23). (B) Whole-cell lysates were western blotted for PTEN, BIM, MYC, and β-actin expression in the noted BL cell lines expressing empty vector (C) or a vector containing mIR17-92 sponge (S). (C) BIM, PTEN, and MYC expression was normalized to β-actin, and relative expression (sponge/vector control) was quantified from 2 separate western blot experiments. Error bars represent 1 standard deviation. (D) Proliferation in BL cell lines after miR17∼92 sponge expression or MIR17HG knock out (Daudi-CRISPR) and/or ibrutinib (IBT) treatment. Line graph depicts 1 representative experiment of triplicate experiments, and points are the average of 3 biological replicates. Error bars represent 1 standard deviation. BL cell lines were treated with a specific concentration of IBT based on the 50% infective dose calculation for each line. (E) Comparison of mRNA levels of CD22 and FCGR2B in control (vector or parental) or miR17∼92 sponge or knockout (Daudi-CRISPR) cells with or without IgM stimulation. Fold change was calculated relative to the vector or parental controls. Graph depicts 1 representative experiment of 3 separate experiments. Error bars depict 1 standard deviation of 3 technical replicates. (F) Cell proliferation in BL cell lines after MYC KO and/or miR17∼92 knockdown (sponge). Points represent the mean of 3 biological replicates from 1 representative experiment of at least 3 separate experiments. Error bars represent the standard error of the mean. (G) Western blots of whole-cell lysates from control (GFP or parental) or miR17∼92 sponge-expressing or MIR17HG knock-out (Daudi-Cas-9) BL cell lines after induction of BCR signaling with anti-IgM. (H) Relative expression of phosphorylated/total SYK, BTK, BLNK, and ERK was quantified from the western blot, suggesting decreased BCR signaling in miR17∼92 sponge-expressing or KO BL cell lines.
We tested whether the absence of MYC (CRISPR-mediated knockout [KO] of MYC) and/or functional loss of miR17∼92 using sponge affected the proliferation and survival of BL cell lines. Sponge effectiveness was assessed using known target genes (PTEN and BIM), and increased expression (>1.25- to 2.5-fold) of target genes was observed, excluding P3HR-1, which does not express PTEN (Figure 6B-C). Consistent with a role in fine-tuning MYC expression,38 a moderate increase in MYC expression was noted in at least 2 cell lines (Figure 6B-C). MiR17∼92-sponge expression led to significantly reduced proliferation (Figure 6D) and G1-S cell cycle arrest in 3 of 4 tested BL cell lines (supplemental Figure 10D). Similar results were obtained with the GA-10 cell line and the Daudi cell line with copy loss of MIR17HG by CRISPR/Cas9 (Figure 6D; supplemental Figure 10E-H). When stressed with reduced serum, miR17∼92 sponge-expressing cells had reduced proliferation (supplemental Figure 10I). Although there was a proliferative disadvantage on miR17∼92 inhibition, only Daudi cells showed an increase in apoptosis (supplemental Figure 10J), possibly as a result of high expression of antiapoptotic proteins (BCLXL and MCL1; supplemental Figure 10C). MYC KO decreased cell proliferation (Figure 6F; supplemental Figure 10K) and increased sensitivity to serum deprivation (supplemental Figure 10L), but miR17∼92 sponge-expressing MYC KO cells showed no major difference in proliferation/viability.
MiR17∼92 expression induces BCR signaling in BL cell lines and can be targeted by ibrutinib
MiR17∼92 inhibits negative regulators of BCR signaling by directly targeting immune-receptor tyrosine-based inhibitory motif-containing genes CD22 and FCGR2B (Figure 6E).40 IgM crosslinking in BL cell lines induced BCR signaling, as measured by kinase activity (phosphorylation) of immediate upstream tyrosine kinase (p-SYK), adaptor protein (p-BLNK), effector protein (p-BTK), and downstream regulators (p-ERK).41 We observed consistently reduced phosphorylation of the above-mentioned BCR regulators in the miR17∼92 sponge-expressing BL cell lines, including GA-10 and the genetically edited Daudi cell line (MIR17HG KO) (Figure 6G-H), suggesting a positive regulation of BCR activation by miR17∼92. The above observation was corroborated by increased mRNA expression of either 1 or both miR17∼92 targets (CD22, FCGR2B) in sponge-expressing BL cell lines (Raji and Daudi).The increase in expression in sponge-expressing cell lines increased significantly on IgM stimulation in 4 BL cell lines (Figure 6E). P3HR1 did not show consistent results. Similar results were observed with 2 other immune-receptor tyrosine-based inhibitory motif-containing negative regulators of BCR signaling, which are direct (FCLR4) or indirect (CD72) targets40 of miR17∼92 (supplemental Figure 10M-N).
FCGR2B recruits SHIP, a phosphoinositol phosphatase, which leads to BTK and PLCγ dissociation and dampens BCR signaling.42 Therefore, we examined the sensitivity of BL cell lines to the BTK inhibitor ibrutinib at their IC50 concentration with and without functional loss of miR17∼92. The BL cell lines were sensitive to ibrutinib in a dose- and time-dependent manner, suggesting dependence on BCR signaling (supplemental Figure 11A-B). However, there was an additive effect of ibrutinib treatment in BL cell lines with miR17∼92 sponge vector compared with control cells, and this was also noted in the P3HR1 cell line. In BL cell lines expressing miR17∼92 sponge, ibrutinib treatment resulted in growth inhibition (Figure 6D). Interestingly, Nalwama cells, which had the highest level of miR17∼92 amplification, were the most sensitive to ibrutinib treatment (supplemental Figure 11B) and were further sensitized in the presence of the miR17∼92 sponge.
Discussion
Reexamination of mBL cases in view of the 2016 World Health Organization classification43 indicated that 8 of 24 adult-mBLs may fit the high-grade B-cell lymphoma category. However, 7 cases had genetic characteristics similar to BL, including MYC, ID3, and TCF3 mutation; del11q, del22q11, low miR-155 expression (CT value ≥30)17 ; and/or CCND3 and TP53 mutations, which are more frequent in BL than other NHLs. These cases had a lower frequency of FL/tFL-characteristic mutations (EZH2, CREBBP), but showed a KMT2D (MLL2) mutation frequency similar to FL (supplemental Figure 1E). These observations suggest that high-grade B-cell lymphomas with BL signature are not derived from FL or GCB-DLBCL, but may have arisen from a precursor that has acquired BCL2 and MYC translocations and, in many cases, KMT2D mutation. The current adult mBL series has dismal outcome, similar to an earlier study (supplemental Figure 1D; data from Sha et al23 ), but different from the adult BL series presented in Dunleavy et al treated with DA-EPOCH-R.44 Suboptimal treatment, patient age (median age 25 years vs 55 years in this series), and diagnostic challenges may contribute to the poor outcome, but a recent meta-analysis of BL across the United States has shown inferior outcome in those older than 40 years, despite improvement in clinical outcome of younger patients.5
The current study showed that BL had fewer CN genomic changes than other GC-derived B-NHLs, indicating that MYC overexpression resulting from t(8;14) is a major driver of lymphomagenesis. This is not surprising, considering that MYC is an amplifier of global transcription,45 regulating 10% to 15% of the transcriptome, especially genes regulating the cell cycle, growth, metabolism, and apoptosis.46 Normally, MYC overexpression induces p53-mediated apoptosis, and tumors overexpressing MYC circumvent this by disrupting the p53 pathway.47 Recurrent CNAs/mutations were identified in genes associated with p53 signaling, cell cycle regulation, or apoptosis, as shown in the results section (supplemental Table 6). Loss of p53 function by CN loss/mutation occurs in half of adult-mBL. TP53 copy loss and cnLOH cases also had TP53 mutation, but not vice versa, indicating mutation may occur earlier than CNA. Other regulators of p53 signaling pathway not previously reported include TS EI24 and TBRG1, which are located within del11q21, a loss unique to mBL. These genes are involved in p53-mediated apoptosis48 and impairment of MDM2-mediated inhibition of p53 transcriptional activity,49 respectively.
Adult-mBLs had more CNAs than pediatric-mBLs, but fewer than other B-cell NHLs. Interestingly, del22q11 is unique to pediatric-BL, and was not found in other adult-mBL cases except 3 double hit cases. Although the loss can be associated with IGL λ locus rearrangement,32 a similar rearrangement in CLL leads to miR-650 upregulation.50 Two other genes in this locus, ZNF280B (33% of pediatric mBL), which positively regulates MDM2 expression,51 and VPREB1 (11% of pediatric mBLs), deleted in pediatric-ALL independent of IGL λ rearrangement,52 are downregulated in cases with copy loss, but their role in BL pathogenesis needs further investigation. Several genetic alterations were either unique (18q21 and 8q24 gain) or more frequent in adult BL (13q31 gain, del13q14, del6q21). Gain of 18q21 encompassed MALT1, BCL2, and TCF4 and had corresponding upregulated mRNA and/or mutation. BCL2 protein/mRNA expression was higher in adult BL, consistent with earlier studies,53 and co-occurred with MCL1 (1q21 gain) another antiapoptotic protein. BL cell lines were more sensitive to a pan-BCL2 inhibitor (Sabutoclax) than to agents targeting single BCL2 family members, suggesting a potential target in adult BL.
mBL had distinct mutations compared with other GC-derived B-NHL, with mutations in genes regulating the cell cycle (MYC, CCND3, TP53; all P ≤ .01 for mBL vs GC-derived B-NHL), the BCR-PI3K pathway (TCF3, ID3), and chromatin modification (SMARAC4) associated with mBL. TCF3 activates PI3K via BCR signaling,27 but also promotes cell proliferation via direct activation of CCND3, essential for expansion of GC B-cells.54 ID3 and TCF3 mutations impair the negative feedback between TCF3 and ID3.27 CCND3 mutations, restricted to the C terminus, a domain crucial for the formation of activating complex with cyclin-dependent kinases (CDK4/CDK6), thus may inhibit interaction with the negative cell cycle regulators p21 and p27.55 CCND3 can inactivate RB1 by phosphorylation, and RB1 deletion/mutations and CCND3 mutations tend not to co-occur, suggesting they regulate a common pathway leading to proliferation.
SMARCA4, an epigenetic regulator, was primarily mutated in pediatric-mBL, whereas KMT2D (MLL2) mutations were mainly in adult-mBL. SMARCA4 loss in B cells can lead to IGH-MYC translocations resulting from impairment of TOP1 recruitment to chromatin,56 and may be a primary lesion in pediatric-BL. Genes that regulate JAK-STAT or chronic BCR-NF-κB signaling and the S1P-pathway were more frequent in adult-mBL and pediatric-mBL respectively. The S1P pathway is frequently disrupted in GC-derived B-NHL, and its disruption impairs the confinement of B cells to the germinal center. Several genes were mutated only in adult-mBL, including the TCF3-related transcription factor TCF4, which was also associated with CN gain and elevated mRNA. TCF4 mutation did not co-occur with TCF3 mutation. Although they share homology and DNA-binding specificity, whether TCF4 and TCF3 functions are redundant needs further investigation.
MYC expression may be regulated by miR17∼92, as it fine tunes MYC translation via Chek2-HuR,38 an observation, noted in at least 2 BL cell lines (supplemental Figure 6C), in which sponge transduction led to a marginal increase in MYC protein expression. Consistent with this model, an inverse correlation between MYC and miR17∼92 expression was observed in BL cases. Furthermore, adult-mBL without MIR17HG gain had higher MYC expression and enrichment of a MYC gene target signature. Together this suggests an oncomiR may substitute for an oncogene in some adult-mBLs.21 MiR17∼92 can modulate the function of multiple pathways promoting proliferation, chemoresistance,21 and BCR signaling40 in B cells. The functional loss of miR17∼92 resulted in reduced proliferation in BL cell lines, which was further enhanced in stress conditions resulting from serum starvation, suggesting miR17∼92 expression is crucial for tumor survival. Loss of miR17∼92 in a cell line lacking expression of PTEN (P3HR1) did not have a significant effect on proliferation unless stressed with reduced serum, suggesting miR17∼92 acts partially through the PI3K pathway. The loss of functional miR17∼92 led to diminished BCR signaling in BL cell lines, consistent with earlier findings in DLBCL cell lines,40 thus demonstrating a positive role in B-cell activation. Because known targets of miR17∼92 include immune-receptor tyrosine-based inhibitory motif-containing genes (CD22, CD72, FCLR4, and FCGR2B), and FCGR2B also modulates BTK function via SHIP and PLCγ,42 we tested the BTK inhibitor approved by the US Food and Drug Administration, ibrutinib, to inhibit BCR signaling in BL lines. Ibrutinib inhibited growth in all BL cell lines in a time- and dose-dependent manner with a range similar to an activated B-cell-DLBCL cell line. The additive growth inhibition of sponge-transduced BL cell lines on ibrutinib treatment suggests dual inhibition of BCR signaling and miR17∼92 by anti-miRs may prove therapeutically synergistic. Our earlier study has shown that BL and HG-BCL cases express elevated levels of miR17∼92 compared with other B-cell lymphomas.17 Now we have shown that MIR17HG focal amplification leads to even higher miR17∼92 expression levels, and thus these patients in particular may benefit from the above therapeutic approaches. However, further evaluation of these inhibitors in vivo is necessary for clinical application, and a more specific BTK inhibitor, such as acalabrutinib, may prove beneficial in adult-BL because of its low toxicity and the ability to transit the blood–brain barrier to reduce central nervous system involvement in BL.
Presented in part at the 58th annual meeting of the American Society of Hematology, San Diego, CA, 5-8 December, 2016.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to acknowledge the Flow Cytometry Research Facility and the Tissue science facility for immunohistochemistry staining at University of Nebraska Medical Center. J.I. was supported in part by the Lymphoma Research Foundation (F-263549), translational research program of Leukemia and Lymphoma Society (6129-14), and National Institutes of Health, National Cancer Institute Eppley Cancer Center Support Grant (P30CA036727). W.C.C. is supported by City of Hope Cancer Center Support Grant (P30CA33572) and Lymphoma SPORE developmental project grant (1 P50 CA 136411-01 01A1 PP-4).
Authorship
Contribution: A.B., C.B., W.L., W.C.C., and J.I. designed and performed the study; W.Z., A.K., T.H., C.M.L., J.Y., R.F., N.E., A. Reddy, T.W.M., and M.G. assisted in research, data analysis, and interpretation; T.C.G., J.V., D.D.W., R.D.G., A. Rosenwald., G.O., E.C., L.M.R., E.S.J., R.M.B., R.S., R.R.M., S.D., J.D., L.M.S., J.Y.S., K.F., and W.C.C. provided materials, reviewed pathology results, or contributed clinical/array comparative genomic hybridization/GEP data; and A.B. and J.I. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Javeed Iqbal, Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, NE 68198-7660; e-mail: jiqbal@unmc.edu; and Wing C. Chan, Department of Pathology, City of Hope Medical Center, Duarte CA, 91010; e-mail: jochan@coh.org.
References
Author notes
A.B., C.B., and W.L. contributed equally to this study.
W.C.C. and J.I. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal