Key Points
Human serum ERFE shows similar pathophysiological responses to mouse models.
Abstract
Erythroferrone (ERFE) is a glycoprotein hormone secreted by erythroblasts in response to stimulation by erythropoietin (EPO). We previously demonstrated that ERFE messenger RNA expression and serum protein concentration increase in mice subjected to hemorrhage or EPO therapy, that ERFE acts on hepatocytes to suppress hepcidin, and that the resulting decrease in hepcidin augments iron delivery for intensified erythropoiesis. We also showed that ERFE contributes to pathological hepcidin suppression and iron overload in mice with nontransfused β-thalassemia. We now report the development and technical validation of a rabbit monoclonal antibody–based sandwich immunoassay for human ERFE. We use this assay to show that blood loss or EPO administration increases serum ERFE concentrations in humans, and that patients with both nontransfused and transfused β-thalassemia have very high serum ERFE levels, which decrease after blood transfusion. The assay should be useful for human studies of normal and disordered erythropoiesis and its effect on iron homeostasis.
Introduction
Intestinal iron absorption and the release of iron from stores increase greatly within hours after blood loss or administration of erythropoietin (EPO).1 In murine models, the response is largely mediated by erythroferrone2 (ERFE, also known as FAM132B), a glycoprotein hormone secreted by EPO-stimulated erythroblasts. ERFE acts by suppressing the hepatic synthesis of the master iron-regulatory hormone, hepcidin.3 Pathologically increased ERFE contributes to hepcidin suppression and iron overload in a mouse model of nontransfused β-thalassemia.4 We report the development of a first-generation assay for human ERFE (hERFE) and show that the assay detects the analogous physiological ERFE increases in humans subjected to blood loss or EPO administration, as well as the pathological increases of ERFE in β-thalassemia.
Study design
Recombinant hERFE production and purification
An hERFE sequence was cloned into pcDNA3.1 with the following modifications: vector signal sequence (interleukin-2) was used instead of the native, followed by a spacer (italics) and a FLAG tag (bolded) (rhERFE1). Because this FLAG tag was mostly lost during cell culture, we further modified the protein by removing the nearby trypsin-sensitive site (strikethrough), which allowed the protein to be secreted efficiently with its FLAG tag (rhERFE2): MYRMQLLSCIALSLALVTNSISAMVRSDYKDDDDKSPEPPPPGNELPRGPGESRAGPAARPPEPTAERAHSVDPRDAWMLFVRQSDKGVNGKKRSRGKAKKLKFGLPGPPGPPGPQGPPGPIIPPEALLKEFQLLLKGAVRQRERAEPEPCTCGPAGPVAASLAPVSATAGEDDDDVVGDVLALLAAPLAPGPRAPRVEAAFLCRLRRDALVERRALHELGVYYLPDAEGAFRRGPGLNLTSGQYRAPVAGFYALAATLHVALGEPPRRGPPRPRDHLRLLICIQSRCQRNASLEAIMGLESSSELFTISVNGVLYLQMGQWTSVFLDNASGCSLTVRSGSHFSAVLLGV.
The antigens and assay standard were produced (see supplemental Data; available on the Blood Web site) in suspension culture in Freestyle 293F cells transiently overexpressing rhERFE1 or rhERFE2. rhERFE1 was purified from supernatant using ion-exchange chromatography. rhERFE2 was purified using an anti-FLAG M2 affinity gel per the manufacturer’s protocol (Sigma). The purified protein was electrophoretically heterogeneous, indicating posttranslational processing and multimerization characteristic of the tumor necrosis factor α–C1q family of proteins.5 Predominant bands on reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis were at 52 kDa and 26 kDa. Antigen concentration was estimated by absorbance (1 mg/mL) at 280 nm = 0.57.
Rabbit monoclonal antibody production
Rabbit hybridomas were generated (Abcam, Burlingame, CA) from rabbits immunized by rhERFE1 and boosted by rhERFE2. Hybridoma supernatants were selected for reactivity against rhERFE2. After biotinylation of monoclonal antibodies (Mabs) (EZ-Link sulfo-NHS-LC-LC-Biotin kit, Thermo Fisher Scientific), optimal pair of unbiotinylated capture Mab and biotinylated detection Mab was chosen by checkerboard testing with rhERFE1 and rhERFE2. The complementary DNAs encoding the final Mab pair (#9 and #42) were cloned from the hybridomas and used to produce the Mabs recombinantly. Peptide epitope scanning showed that Mab #9 bound to the peptide ELPRGPGESRAGPAARPP but not to 6 amino acid overlap neighbors suggesting that it was specific for an epitope centered on the underlined segment GESRAG. Mab #42 did not bind to linear peptides, indicating that it probably recognized a 3-dimensional epitope.
hERFE immunoassay (see supplemental Data for details)
The 96-well plates were coated with Mab #9, washed, and blocked. Recombinant hERFE2 standard was serially diluted to 10, 5, 2.5, 1.25, and 0.625 ng/mL. After a 1 hour incubation, the plate was washed and incubated for 1 hour with 100 µL per well biotinylated Mab #42 (1 µg/mL). The plate was then washed, incubated for 45 minutes with Neutravidin–horseradish peroxidase conjugate 1/5000 (100 µL per well), and developed with tetramethylbenzidine Substrate System for enzyme-linked immunosorbent assay (ThermoScientific) at room temperature for 10 minutes. The reaction was stopped by adding 50 µL of 2N sulfuric acid, and the plates were read on a Spectramax 250 (Molecular Devices) at 450 nm.
Hepcidin assay
Hepcidin was measured by competitive enzyme-linked immunosorbent assay6 (Intrinsic Life Sciences, La Jolla, CA).
Human samples
All human studies were approved by institutional review boards at respective institutions and at University of California, Los Angeles (UCLA). All samples were venous sera.
Blood donors.
Male blood donors at the New York Blood Center (n = 30, age 19-65 years) donated 2 units of packed erythrocytes by apheresis (see supplemental Data for details). Sera were collected prior to and 2, 4, 7, 9, 11, 14, and 112 days following donation. Sera from female blood donors (n = 30, age 18-61) were obtained from Discovery Life Sciences (San Luis Obispo, CA).
EPO administration.
Four geriatric patients at UCLA with moderate anemia (hemoglobin 9.5-10.3 g/dL) of unknown etiology were administered 20 000 units of EPO subcutaneously, and sera collected over a 1-week time course.
β-thalassemia.
Patients were recruited at the UCSF Benioff Children's Hospital Oakland and included nontransfused (n = 11, 10 male [M], 1 female [F], age range 6-69 years) or transfusion-dependent patients immediately before (n = 10, 4 M, 6 F, 6-38 years) or 2 to 14 days after transfusion (n = 13, 9 M, 4 F, 5-36 years).
Results and discussion
Properties of the ERFE assay
The standard curve was linear after log-log transformation. Limit of blanks (64 replicates), calculated as average + 1.645 × standard deviation (SD) of blanks, was 0.8 ng/mL. The limit of detection, calculated as average + 1.645 × SD of the concentration calculated from 64 replicates of the lowest standard (0.625 ng/mL), was 1.5 ng/mL. The lower limit of quantitation was determined as 14 ng/mL by analyzing at 10-fold sample dilution, the coefficient of variation percent (CV%) of 16 replicates each of 8 human samples with low ERFE concentrations, graphing CV% vs ERFE concentration, fitting the relationship with an exponential curve, and interpolating an ERFE concentration that yielded CV% = 20. For 4 of these samples ranging from 17 to 25 ng/mL, the CV% was 6.8 ± 0.8. The working range was therefore 14 to 100 ng/mL. Spike recovery was determined by adding 2.5, 5.0, or 10 ng/mL of hERFE2 to 10-fold dilutions of human serum samples (n = 9) containing very low concentrations of ERFE (0-0.8 ng/mL), measuring for each sample the resulting ERFE concentration and subtracting its prespike ERFE concentration. The spike recovery (mean ± SD) was 92 ± 8%, 100 ± 5%, and 111 ± 4% for spikes of 2.5, 5.0, or 10 ng/mL respectively, corresponding to sample concentrations of 25, 50, and 100 ng/mL. The performance of the assay in repeat testing of sera stored at 4°C or frozen at −80°C for up to 28 days is shown in supplemental Data and supplemental Figure 1.
ERFE response to erythropoietic stimulation
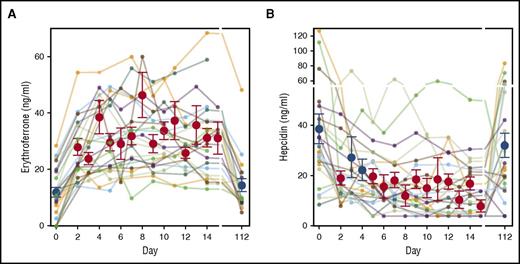
ERFE concentration in male, female, and combined blood donors prior to donation (baseline) was 12 ± 9 ng/mL (mean ± SD, n = 28), 11 ± 11 ng/mL (n = 30), and 12 ± 10 ng/mL (n = 58). The distributions were skewed so that median and percentile range (25%, 75%) was 12 (7, 19), 7 (4, 9), and 8 (4, 15) for men, women, and combined genders. Follow-up on the male donors for up to 112 days (Figure 1A) showed that serum ERFE rose in the second sample in all donors (on average 2.5 days after donation (range 2-4 days) to 28 ± 11 ng/mL (P = 3 × 10−7, paired Student t test compared with baseline). Further rise of serum ERFE was seen in most donors with a maximum reaching 38 ± 13 ng/mL (n = 29, P = 5 × 10−11) at 9 ± 4 days. At 112 days following donation, serum ERFE concentrations returned to baseline (15 ± 10 ng/mL, P = .3, n = 23). Mean serum hepcidin was suppressed on days 2 to 15 but returned toward baseline by 112 days (Figure 1B).
Serum ERFE and hepcidin after blood donation. (A) Serum ERFE in blood donors of 2 units of erythrocytes. Colored lines denote individual donors. Red and blue circles and error bars show mean ± standard error of the mean for each time point. Red denotes P < .05 by 1-way repeated measures analysis of variance (ANOVA) compared with initial baseline (day 0) concentration. The first postdonation sample was obtained at average 2.5 days after donation (range 2-4 days) and showed an increase in all samples, by 16.4 ± 8.9 ng/mL over initial sample (mean increase ± SD, P = .0002, paired Student t test, n = 24). Further rise of serum ERFE was seen in most donors with a maximum at 8 ± 4 days (mean ± SD, n = 24) increasing by 26.7 ± 11.2 ng/mL over initial value. By 112 days after the blood donation, serum ERFE concentrations returned to baseline (3.9 ± 7.5 ng/mL over initial, P = .28 comparing baseline and 112 days, paired Student t test, n = 20). (B) Serum hepcidin in the same donors is shown for reference. Red denotes P < .05 by 1-way repeated measures ANOVA compared with initial baseline (day 0) concentration.
Serum ERFE and hepcidin after blood donation. (A) Serum ERFE in blood donors of 2 units of erythrocytes. Colored lines denote individual donors. Red and blue circles and error bars show mean ± standard error of the mean for each time point. Red denotes P < .05 by 1-way repeated measures analysis of variance (ANOVA) compared with initial baseline (day 0) concentration. The first postdonation sample was obtained at average 2.5 days after donation (range 2-4 days) and showed an increase in all samples, by 16.4 ± 8.9 ng/mL over initial sample (mean increase ± SD, P = .0002, paired Student t test, n = 24). Further rise of serum ERFE was seen in most donors with a maximum at 8 ± 4 days (mean ± SD, n = 24) increasing by 26.7 ± 11.2 ng/mL over initial value. By 112 days after the blood donation, serum ERFE concentrations returned to baseline (3.9 ± 7.5 ng/mL over initial, P = .28 comparing baseline and 112 days, paired Student t test, n = 20). (B) Serum hepcidin in the same donors is shown for reference. Red denotes P < .05 by 1-way repeated measures ANOVA compared with initial baseline (day 0) concentration.
We also examined the response to the administration of EPO (20 000 units) in 4 geriatric patients with moderate anemia of unknown etiology (Figure 2A-B). ERFE increased in all patients, reaching a maximum at 1 to 5 days, coincident with a decline in serum hepcidin concentrations.
Serum ERFE and hepcidin in EPO-treated patients and in β-thalassemia. (A) Serum ERFE in 4 patients with moderate anemia treated on day 0 with EPO 20 000 units subcutaneously. Blood samples were obtained every 6 hours the first day, then on mornings of days 2, 3, 5, and 7. (B) Serum hepcidin in the same samples as panel A. The patient designated by green symbols was iron deficient (serum ferritin 14, transferrin saturation 17%, undetectable serum hepcidin). (C) Serum ERFE in patients with β-thalassemia, nontransfused (Non), before transfusion (Pre), or after transfusion (Post). Box plots show median, box 25% to 75%, whiskers 10% to 90%, and outliers. Serum ERFE levels were massively increased in nontransfused and pretransfusion thalassemic patients but were closer to normal after transfusion, *P < .05, 1-way ANOVA on ranks, comparing with normal reference group of combined male and female blood donors before donation. (D) Serum hepcidin in the same samples, vs ERFE; scatterplot is fitted by an inverse third-order equation, R2 = 0.7 and P < .0001.
Serum ERFE and hepcidin in EPO-treated patients and in β-thalassemia. (A) Serum ERFE in 4 patients with moderate anemia treated on day 0 with EPO 20 000 units subcutaneously. Blood samples were obtained every 6 hours the first day, then on mornings of days 2, 3, 5, and 7. (B) Serum hepcidin in the same samples as panel A. The patient designated by green symbols was iron deficient (serum ferritin 14, transferrin saturation 17%, undetectable serum hepcidin). (C) Serum ERFE in patients with β-thalassemia, nontransfused (Non), before transfusion (Pre), or after transfusion (Post). Box plots show median, box 25% to 75%, whiskers 10% to 90%, and outliers. Serum ERFE levels were massively increased in nontransfused and pretransfusion thalassemic patients but were closer to normal after transfusion, *P < .05, 1-way ANOVA on ranks, comparing with normal reference group of combined male and female blood donors before donation. (D) Serum hepcidin in the same samples, vs ERFE; scatterplot is fitted by an inverse third-order equation, R2 = 0.7 and P < .0001.
Pathological increase of ERFE in β-thalassemia
ERFE concentrations in nontransfused and pretransfusion patients were greatly increased compared with our reference sample from blood donors at baseline (Figure 2C; P < .05, 1-way ANOVA on ranks, Dunn’s method). Hepcidin concentrations inversely correlated with ERFE concentrations, consistent with the proposed role of ERFE as a pathological hepcidin suppressor in β-thalassemia (Figure 2D; third-order equation, R2 = 0.7, P < .001).
Conclusion
The assay for hERFE should be useful in understanding the pathophysiological interaction between erythropoiesis and iron homeostasis including the pathogenesis of iron-loading anemias, erythropoietic response to therapy with erythropoiesis-stimulating agents in chronic kidney disease, anemia of cancer or anemia of inflammation, and physiological adaptations to hypoxia, altitude, or blood donation. Future tasks include standardization of the reference antigen and the development of more reliable laboratory reference ranges in healthy adults and children. Further improvement in assay performance can be expected with automation and close control of environmental variables.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Virginia Donovan (Abcam) for producing the rabbit monoclonal antibodies and many collegial interactions during the process, and Erika Valore (UCLA and Stanford) for her advice and guidance.
This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (R01 DK 065029) (T.G.), and by the UCLA Center for Accelerated Innovation, under National Heart, Lung, and Blood Institute, National Institutes of Health (UC CAI grant U54HL119893) (M. Palazzolo, Principal Investigator) and National Center for Advancing Translational Sciences, National Institutes of Health (UCLA CTSI grant UL1TR001881).
Authorship
Contribution: T.G. conceived the study, obtained funding, directed the study, analyzed the data and wrote the manuscript; G.J. performed experiments and analyzed the data; A.N. organized human studies and edited the manuscript; Y.G. organized human studies and edited the manuscript; Z.P. organized human studies and recruited patients; P.B.W. organized human studies and edited the manuscript; L.K. designed and performed experiments and edited the manuscript; E.N. codirected the study, analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: T.G., L.K., and E.N. are inventors on a patent application on ERFE. T.G. and E.N. are scientific founders of Intrinsic LifeSciences and Silarus Pharma, companies that have interests related to ERFE.
Correspondence: Tomas Ganz, David Geffen School of Medicine at University of California Los Angeles, 10833 Le Conte Ave, CHS 52-243, Los Angeles, CA 90095-1690; e-mail: tganz@mednet.ucla.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal