To the editor:
Recent advances in understanding the contributions of the contact pathway to thrombosis, inflammation, and innate immunity1 have renewed interest in identifying (patho)physiologic activators of this pathway. Candidates include collagen,2 misfolded proteins,3 DNA,4 RNA,4 and polyphosphate (polyP).5 Contributions of extracellular nucleic acids to coagulation and inflammation are under intense investigation, with several studies reporting the procoagulant effects of DNA,6-8 particularly in the context of neutrophil extracellular traps (NETs).9-11 In this study, we attempted to compare the clotting activity of polyP versus cellular DNA. To do this, we isolated DNA from human cells using Qiagen kits, as previously done by others (Table 1). We now report that DNA purified by using these kits was contaminated with highly procoagulant silica particles. Some studies have employed Qiagen kits to isolate polyP from human cells (Table 1), and we also now report that polyP isolated by using such kits risks contamination with procoagulant silica.
Nucleic acid purification methods used in published studies of the procoagulant activities of polyP or DNA
| Kit name (Manufacturer) . | Principle . | Used to isolate . | Reference . |
|---|---|---|---|
| Qiaquick PCR Purification (Qiagen) | Silica | polyP | 12 |
| 13 | |||
| DNeasy Blood and Tissue (Qiagen) | Silica | DNA | 14 |
| 8 | |||
| QIAamp Blood (Qiagen) | Silica | DNA | 10 |
| 7 | |||
| 15 | |||
| 16 | |||
| Genomic DNA isolation kit (Qiagen) | Silica | DNA | 4 |
| DNAzol (Invitrogen) | Guanidinium isothiocyanate/ detergent | DNA | 17 |
| — | Phenol/chloroform extraction | DNA | 18 |
| 19 |
| Kit name (Manufacturer) . | Principle . | Used to isolate . | Reference . |
|---|---|---|---|
| Qiaquick PCR Purification (Qiagen) | Silica | polyP | 12 |
| 13 | |||
| DNeasy Blood and Tissue (Qiagen) | Silica | DNA | 14 |
| 8 | |||
| QIAamp Blood (Qiagen) | Silica | DNA | 10 |
| 7 | |||
| 15 | |||
| 16 | |||
| Genomic DNA isolation kit (Qiagen) | Silica | DNA | 4 |
| DNAzol (Invitrogen) | Guanidinium isothiocyanate/ detergent | DNA | 17 |
| — | Phenol/chloroform extraction | DNA | 18 |
| 19 |
Detailed methods used to purify and analyze DNA and polyP are described in the supplemental Methods. Briefly, DNA was purified from HEK 293 cells or human NETs by using DNeasy Blood and Tissue kits (Qiagen) or phenol/chloroform extraction. Some preparations of λ phage DNA or short-chain polyP were repurified on DNeasy kits. Water elutions from DNeasy kits or Econospin (Epoch Life Sciences, Missouri City, TX) columns were prepared by skipping the loading and washing steps and substituting water for the elution buffer. Some samples were digested with calf intestinal alkaline phosphatase (a potent exopolyphosphatase) and Benzonase (a nonspecific nuclease). Other samples were boiled in 1 M hydrochloric acid, and then neutralized and buffer-exchanged into clotting buffer. Digestion of DNA or polyP was confirmed by electrophoresis (supplemental Figure 1).
DNA purified from NETs or HEK 293 cells by using Qiagen DNeasy kits exhibited robust procoagulant activities (Figure 1A), albeit with batch-to-batch variability (supplemental Figure 2). In contrast, DNA purified from these same cells by using phenol/chloroform extraction exhibited limited procoagulant activity (Figure 1A; supplemental Figure 2). Thus, Qiagen DNeasy kits somehow rendered the purified DNA much more procoagulant.
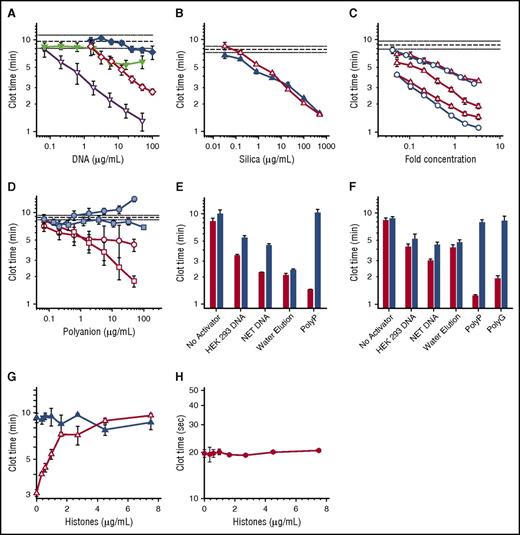
Procoagulant activities of DNA, polyP, and silica particles. (A-D) Shown are plasma clot times versus concentration, with horizontal dashed lines showing the mean clot time without activator (± standard error of the mean as horizontal dotted lines). (A) Clot times with: HEK 293 cell DNA isolated with DNeasy Blood & Tissue (open diamond); HEK 293 cell DNA isolated with phenol/chloroform (solid diamond); NET-derived DNA isolated with DNeasy Blood & Tissue kit (open inverted triangle); or NET-derived DNA isolated with phenol/chloroform (solid inverted triangle). Each data set represents the mean clot time for 3 separate purifications as detailed in supplemental Figure 2A (HEK293 cell DNA) and supplemental Figure 2B (NET DNA). (B) Clot times with silica particles: glass milk (solid triangle) or homogenized DNeasy column matrix (open triangle). (C) Clot times with water elutions from 3 different lots of DNeasy columns (open triangle) or 2 different lots of Econospin columns (open circle). On the x-axis, fold concentration refers to the concentration relative to the volume eluted from the Qiagen column, with 1 equaling the original elution volume. (D) Clot times of λ phage DNA before (blue square) or after (open square) repurification on DNeasy columns; or of short-chain polyP before (blue circle) or after (open circle) repurification on DNeasy columns. The purification data sets represent mean clot times for 2 different purifications as detailed in supplemental Figure 4. (E) Clot times of various samples before (red bars) or after (blue bars) digestion with a combination of Benzonase and calf intestine alkaline phosphatase. Samples include buffer control (no activator), 20 µg/mL HEK 293 cell DNA purified using DNeasy, 20 µg/mL NET DNA purified using DNeasy, water applied to DNeasy column as described in panel C (water elution), or 2 µg/mL long-chain polyP. Digestion of DNA by Benzonase and polyP by phosphatase was confirmed by gel electrophoresis (supplemental Figure 1). (F) Clot times of various samples before (red bars) or after (blue bars) acid hydrolysis. Samples include buffer control (no activator), 20 µg/mL HEK 293 cell DNA purified using DNeasy, 20 µg/mL NET DNA purified using DNeasy, water applied to DNeasy column (water elution), 2 µg/mL long-chain polyP, or 5 µg/mL polyguanylate (polyG). (G) Clot times in the presence of varying concentrations of histones with (open triangle) or without (solid triangle) a 10-fold concentrated water elution (prepared as described for panel C). (H) Tissue factor–triggered clot times in the presence of varying concentrations of histones.
Procoagulant activities of DNA, polyP, and silica particles. (A-D) Shown are plasma clot times versus concentration, with horizontal dashed lines showing the mean clot time without activator (± standard error of the mean as horizontal dotted lines). (A) Clot times with: HEK 293 cell DNA isolated with DNeasy Blood & Tissue (open diamond); HEK 293 cell DNA isolated with phenol/chloroform (solid diamond); NET-derived DNA isolated with DNeasy Blood & Tissue kit (open inverted triangle); or NET-derived DNA isolated with phenol/chloroform (solid inverted triangle). Each data set represents the mean clot time for 3 separate purifications as detailed in supplemental Figure 2A (HEK293 cell DNA) and supplemental Figure 2B (NET DNA). (B) Clot times with silica particles: glass milk (solid triangle) or homogenized DNeasy column matrix (open triangle). (C) Clot times with water elutions from 3 different lots of DNeasy columns (open triangle) or 2 different lots of Econospin columns (open circle). On the x-axis, fold concentration refers to the concentration relative to the volume eluted from the Qiagen column, with 1 equaling the original elution volume. (D) Clot times of λ phage DNA before (blue square) or after (open square) repurification on DNeasy columns; or of short-chain polyP before (blue circle) or after (open circle) repurification on DNeasy columns. The purification data sets represent mean clot times for 2 different purifications as detailed in supplemental Figure 4. (E) Clot times of various samples before (red bars) or after (blue bars) digestion with a combination of Benzonase and calf intestine alkaline phosphatase. Samples include buffer control (no activator), 20 µg/mL HEK 293 cell DNA purified using DNeasy, 20 µg/mL NET DNA purified using DNeasy, water applied to DNeasy column as described in panel C (water elution), or 2 µg/mL long-chain polyP. Digestion of DNA by Benzonase and polyP by phosphatase was confirmed by gel electrophoresis (supplemental Figure 1). (F) Clot times of various samples before (red bars) or after (blue bars) acid hydrolysis. Samples include buffer control (no activator), 20 µg/mL HEK 293 cell DNA purified using DNeasy, 20 µg/mL NET DNA purified using DNeasy, water applied to DNeasy column (water elution), 2 µg/mL long-chain polyP, or 5 µg/mL polyguanylate (polyG). (G) Clot times in the presence of varying concentrations of histones with (open triangle) or without (solid triangle) a 10-fold concentrated water elution (prepared as described for panel C). (H) Tissue factor–triggered clot times in the presence of varying concentrations of histones.
Qiagen kits for purifying nucleic acids employ spin columns with a silica matrix on which the DNA is adsorbed. Because silica is potently procoagulant,20,21 we tested the hypothesis that Qiagen DNeasy spin columns leach silica particles that contribute to the observed procoagulant activity of DNA. We confirmed the procoagulant activity of silica particles (glass milk) and found that the DNeasy column matrix material was similarly procoagulant (Figure 1B). We next tested our hypothesis by performing mock purifications of phosphate buffered saline (without cells). The eluted material was indeed procoagulant (supplemental Figure 3). We then loaded water onto silica columns from 2 manufacturers. The flow-through was also procoagulant (Figure 1C), with variable activity from different column lots. Elemental analysis of the water elutions from DNeasy columns detected silicon at 5.6 to 98.3 µg/mL, expressed as silicon dioxide (supplemental Table 1).
Some studies have used Qiagen kits to purify polyP from cells (Table 1). We confirmed22 that synthetic, short-chain polyP had a negligible ability to trigger clotting, as did purified bacteriophage λ DNA (Figure 1D). When repurified using DNeasy kits, both λ DNA (Figure 1D; supplemental Figure 4A) and short-chain polyP (Figure 1D; supplemental Figure 4B) acquired significant procoagulant activity. When ethanol precipitation of the eluted λ DNA was substituted for the buffer-exchange/concentration procedure, the DNA still contained procoagulant activity (supplemental Figure 5). This strongly suggested that the kits contribute procoagulant material to the eluted products, and that this material cannot be removed from kit-purified DNA by ethanol precipitation. We attempted to remove this procoagulant material from kit-purified DNA using filtration and centrifugation, but were unable to identify conditions that did not also result in severe DNA loss (data not shown). Some of the contaminating silica particles may be so small that they cannot readily be removed from DNA solutions by using filtration or centrifugation.
We conducted experiments to identify how much of the procoagulant activity in DNA isolated by using DNeasy kits was attributable to DNA versus contaminating silica. Accordingly, we digested the DNA preparations with Benzonase and alkaline phosphatase (in case any polyP had copurified with the cellular DNA). Although electrophoresis confirmed complete degradation of DNA and polyP (supplemental Figure 1), this treatment only partially neutralized the procoagulant activity of cell-derived DNA (Figure 1E). Enzymatic digestion had no effect on the clotting activity of water elution from DNeasy columns, but destroyed the clotting activity of long-chain polyP (Figure 1E). We also subjected aliquots of these DNA preparations to acid hydrolysis (Figure 1F). Although the procoagulant activities of long-chain polyP and a synthetic RNA (polyG) were completely eliminated by heating in acid, significant amounts of procoagulant activity in the Qiagen kit–purified DNA samples survived. The clotting activity of water elution from Qiagen kits was unaffected by acid hydrolysis (Figure 1F).
Noubouossie et al16 demonstrated that the procoagulant activity of Qiagen kit-purified cellular DNA was eliminated by adding histones, proposing that DNA’s procoagulant activity is masked when assembled into nucleosomes. We hypothesized that positively-charged histones might neutralize the procoagulant activity of contaminating silica particles. Indeed, histones completely abrogated the procoagulant activity of “water elutions” (Figure 1G), but had no effect on tissue factor-initiated clotting (Figure 1H). An alternative explanation for the findings of Noubouossie et al is that DNA purified from NETs gained procoagulant activity not because of removal of the histones, but because of silica contamination.
Verhoef et al13 recently reported that polyP isolated from human platelets by using Qiagen PCR Purification kits triggered clotting via the contact pathway, whereas platelet polyP isolated by using phenol/chloroform extraction had little procoagulant activity. To account for these markedly different activities, these authors proposed that the 2 methods isolated polyP of different polymer lengths. A plausible alternative is that platelet polyP isolated by using Qiagen kits was contaminated with highly procoagulant silica, whereas the phenol/chloroform-purified polyP was not.
We found that DNA isolated from either HEK 293 cells or NETs by using phenol/chloroform extraction exhibited measurable ability to shorten plasma clot times, albeit with considerably less procoagulant activity than when purified by using Qiagen kits. To rule out the possibility that carryover of contaminants somehow interfered with the procoagulant activity of DNA, we added phenol/chloroform-purified HEK 293 cell DNA to silica particles eluted from Qiagen kits, and found no significant diminution of the procoagulant activity of this material (supplemental Figure 6). Other groups who purified cellular DNA using non–silica-based methods have also reported measurable ability of cellular DNA to activate the contact pathway. Oehmcke et al17 found that neutrophil-derived DNA (isolated by using DNAzol) bound and activated contact factors. Kokoye et al19 reported that genomic DNA isolated from leukocytes (by using phenol/chloroform extraction) promoted factor XII activation, whereas Ivanov et al23 described the ability of NET-derived DNA (purified by using phenol/chloroform extraction) to support factor XI activation and enhance thrombin generation.
Our results indicate that much, but not all, of the apparent procoagulant activity of cellular DNA isolated by using Qiagen kits may be attributable to traces of highly procoagulant silica particles. In addition, the amount of procoagulant particles shed into DNA preparations is highly variable. Thus, isolating nucleic acids by using silica-based kits risks contamination with silica that could confound the interpretation of experiments on their procoagulant properties. We strongly recommend that studies of the clotting activities of nucleic acids or polyP avoid silica-based methods.
Authorship
Acknowledgments: The authors thank Rachel Hemp and Yuqi Wang for their assistance with cell culture and material preparation and Kiran Subedi (University of Illinois Microanalysis Laboratory) for performing the inductively coupled plasma spectrometry.
This work was supported by grants R01 HL047014, R35 HL135823, and UM1 HL120877 from the National Institutes of Health, National Heart, Lung and Blood Institute.
Contribution: S.A.S., C.J.B., and J.M.G. designed and performed experiments, analyzed results, and edited the manuscript; and J.H.M. designed experiments, analyzed results, and edited the manuscript.
Conflict of interest disclosure: J.H.M. and S.A.S. are coinventors on patents and pending patent applications related to medical uses of polyP. The remaining authors declare no competing financial interests.
The current affiliation for J.M.G. is L.E.K. Consulting, Boston, MA.
Correspondence: James H. Morrissey, Department of Biochemistry, University of Illinois at Urbana-Champaign, 323 Roger Adams Laboratory, MC-712, 600 S Mathews Ave, Urbana, IL 61801; e-mail: jhmorris@illinois.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal