Key Points
The presence of Smad1 or Smad5 in hepatocytes is sufficient to maintain iron homeostasis, whereas deficiency of both induces iron overload.
Erythropoietin and erythroferrone fail to suppress hepcidin in mice with a conditional ablation of Smad1 and Smad5 in hepatocytes.
Abstract
Anemia suppresses liver hepcidin expression to supply adequate iron for erythropoiesis. Erythroferrone mediates hepcidin suppression by anemia, but its mechanism of action remains uncertain. The bone morphogenetic protein (BMP)–SMAD signaling pathway has a central role in hepcidin transcriptional regulation. Here, we explored the contribution of individual receptor-activated SMADs in hepcidin regulation and their involvement in erythroferrone suppression of hepcidin. In Hep3B cells, SMAD5 or SMAD1 but not SMAD8, knockdown inhibited hepcidin (HAMP) messenger RNA (mRNA) expression. Hepatocyte-specific double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice exhibited ∼90% transferrin saturation and massive liver iron overload, whereas Smad1fl/fl;Smad5fl/wt;Cre+ mice or Smad1fl/wt;Smad5fl/fl;Cre+ female mice with 1 functional Smad5 or Smad1 allele had modestly increased serum and liver iron, and single-knockout Smad5fl/fl;Cre+ or Smad1fl/fl;Cre+ mice had minimal to no iron loading, suggesting a gene dosage effect. Hamp mRNA was reduced in all Cre+ mouse livers at 12 days and in all Cre+ primary hepatocytes. However, only double-knockout mice continued to exhibit low liver Hamp at 8 weeks and failed to induce Hamp in response to Bmp6 in primary hepatocyte cultures. Epoetin alfa (EPO) robustly induced bone marrow erythroferrone (Fam132b) mRNA in control and Smad1fl/fl;Smad5fl/fl;Cre+ mice but suppressed hepcidin only in control mice. Likewise, erythroferrone failed to decrease Hamp mRNA in Smad1fl/fl;Smad5fl/fl;Cre+ primary hepatocytes and SMAD1/SMAD5 knockdown Hep3B cells. EPO and erythroferrone reduced liver Smad1/5 phosphorylation in parallel with Hamp mRNA in control mice and Hep3B cells. Thus, Smad1 and Smad5 have overlapping functions to govern hepcidin transcription. Moreover, erythropoietin and erythroferrone target Smad1/5 signaling and require Smad1/5 to suppress hepcidin expression.
Introduction
Iron is an essential nutrient that participates in numerous enzymatic reactions and biological functions; however, too much iron can be toxic because of free-radical generation.1 Abnormalities in systemic iron homeostasis affect nearly 1 billion people worldwide, leading to diseases such as anemia and hemochromatosis.2,3 Hepcidin is a peptide hormone secreted by the liver that plays a central role in regulating systemic iron balance by promoting degradation of the iron exporter ferroportin to inhibit dietary iron absorption and iron recycling from body stores.4 Hepcidin expression is induced by iron as a negative feedback mechanism to maintain steady-state iron levels5,6 and by inflammation to limit iron availability to pathogenic microorganisms.7,8 Conversely, hepcidin expression is inhibited by anemia and other stimulators of erythropoietic drive to increase the iron supply for erythropoiesis.9
At the molecular level, the bone morphogenetic protein (BMP)–SMAD signaling pathway is a major transcriptional regulator of hepcidin. Not only is BMP-SMAD signaling central to hepcidin regulation by iron,10,11 but it also intersects with most other known hepcidin regulators.12 One of the least well-understood pathways is how hepcidin expression is suppressed by erythropoietic drive. Erythropoietic suppression of hepcidin is dependent on an intact bone marrow and occurs indirectly through secreted factor(s) produced by proliferating red blood cell precursors.13 Recently, erythroferrone was demonstrated to be one such mediator of hepcidin suppression by erythropoietic drive.14 How erythroferrone suppresses hepcidin production and whether this pathway intersects with the BMP-SMAD signaling pathway remain uncertain.
BMPs act by binding to a complex of type I and type II serine/threonine kinase receptors to induce the phosphorylation of receptor-activated SMAD transcription factors (R-SMADs), which translocate to the nucleus to modulate gene expression after complexing with SMAD4.15 Genetic mouse models have yielded important insights into the specific components of the BMP signaling pathway that control hepcidin production. These data suggest a model where the ligands BMP6 and BMP2 are produced in liver endothelial cells and have paracrine actions on BMP receptors and the coreceptor hemojuvelin in hepatocytes to regulate hepcidin transcription. Indeed, global or endothelial knockout of Bmp6,16,17 endothelial knockout of Bmp2,18 and global or hepatocyte knockout of Hfe2 (encoding hemojuvelin)10,19 in mice each lead to hepcidin deficiency and iron overload. Hepatocyte knockout of the BMP type I receptors Acvr1 or Bmpr1a also lead to hepcidin deficiency and iron overload, suggesting that both type I receptors are essential for hepcidin regulation.20 For type II receptors, hepcidin expression and iron homeostasis are only impaired in mice that lack both Acvr2a and Bmpr2 in hepatocytes, suggesting that these type II receptors have redundant functions in hepcidin regulation.21
Although the contributions of specific BMP ligands and receptors have been well established, little is known about the relative contribution of R-SMADs in hepcidin regulation and iron homeostasis. Three R-SMADs are phosphorylated in response to BMP signals: SMAD1, SMAD5, and SMAD8 (also known as SMAD9). Liver SMAD1/5/8 phosphorylation is induced by iron in parallel with hepcidin,22 and SMAD signaling is critical to BMP regulation of hepcidin expression because specific SMAD binding elements on the hepcidin promoter are required for hepcidin induction by BMPs.23 Moreover, hepatocyte knockout of common mediator Smad4 leads to hepcidin deficiency and iron overload.24 Although R-SMADs have been shown to have redundant, dose-dependent functions in many biological contexts,25-27 they do not always have overlapping functions. For example, in zebrafish embryos, Smad1 knockdown impairs myelopoiesis but enhances erythropoiesis, whereas Smad5 knockdown causes erythropoiesis failure but normal macrophage numbers.28 Additionally, transcript profiling shows a large set of genes that are regulated independently by Smad1 and Smad5.28 Global Smad1 or Smad5 knockout mice are embryonic lethal, but for different reasons, whereas Smad8 knockout mice are viable.25,29-33 Although some differences can be explained by when, where, and to what extent R-SMADs are expressed, R-SMADs may also bind to different elements and transcriptional coactivators.33-39
Here, we used in vitro studies and conditional knockout mice to determine the relative contribution of R-SMADs to hepcidin regulation and iron homeostasis. We also took advantage of these mouse models to investigate whether the suppression of hepcidin by erythropoietic drive involves the BMP-SMAD pathway.
Methods
Cell culture and transfections
Human hepatoma Hep3B cells were cultured as previously described10 and reverse transfected with 20-nM small interfering RNA(siRNA) targeting SMAD1, SMAD5, SMAD8, or Control siRNA (Dharmacon) using DharmaFECT 4 (Dharmacon) for 48 hours. Primary hepatocytes were isolated and cultured as described in the supplemental Methods (available on the Blood Web site). Where indicated, cells were serum starved overnight with 1% fetal bovine serum and stimulated with Bmp6 (R&D Systems) at 5 ng/mL for 6 hours or treated with conditioned medium containing 50% (volume-to-volume ratio) cell supernatant from control HEK293T cells or HEK293T cells overexpressing erythroferrone (Erfe-CM)14 for 15 or 6 hours.
Animals
Mice harboring LoxP-flanked alleles of both Smad1 and Smad5 (Smad1fl/fl;Smad5fl/fl) on a mixed C57BL/6J;129S5/SvEvBrd background26 were crossed with mice expressing a Cre transgene under the control of a hepatocyte-specific albumin promoter40 on a C57BL/6J background (Jackson Laboratory). Smad1fl/+;Smad5fl/+;Cre+ offspring were either backcrossed with Smad1fl/fl;Smad5fl/fl mice to generate Smad1fl/fl;Smad5fl/fl;Cre+, Smad1fl/wt;Smad5fl/fl;Cre+, and Smad1fl/fl;Smad5fl/wt;Cre+ mice or intercrossed with Smad1fl/+;Smad5fl/+;Cre− to generate single-knockout Smad5fl/fl;Cre+ and Smad1fl/fl;Cre+ mice. Cre+ mice were compared with littermate Cre− controls throughout the study. Mice were weaned at 3 weeks, maintained on a standard diet (Prolab RMH 3000; LabDiet) containing 380 ppm iron, and genotyped as previously described.26,41 Where indicated, mice were treated with a low-iron (2-6 ppm) diet (TD.80396; Harlan Laboratories), epoietin alfa (EPO; 200 U per mouse; Amgen), or neutralizing BMP6 (5 mg/kg of body weight; MAB507; R&D Systems)16 and BMP2/4 antibodies (10 mg/kg of body weight; MAB3552; R&D Systems; supplemental Figure 1). Animal protocols were approved by the Institutional Animal Care and Use Committee at Massachusetts General Hospital.
RNA extraction and quantitative reverse-transcriptase PCR
Total RNA was isolated using Qiashredder and RNeasy Mini Kit (Qiagen). First-strand complementary DNA (cDNA) was synthesized from 1 μg of RNA using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). Polymerase chain reactions (PCRs) were performed using the PowerUp SYBR Green Master Mix on the QuantStudio3 real-time PCR system (Applied Biosystems) using primers listed in supplemental Table 1. Transcript levels were normalized to Rpl19 as an internal control. Transcript copy numbers of SMAD1 and SMAD5 were determined using Taqman Universal Master Mix, TaqMan primers and probes that were selected to target all known mRNA variants (Applied Biosystems), and standard curves that were generated from the plasmids listed in supplemental Table 2.
Iron analysis
Serum iron and unsaturated iron binding capacity were measured by colorimetric assay (Pointe Scientific) to calculate transferrin saturation according to manufacturer’s instructions. Tissue nonheme iron concentrations (in micrograms per gram wet weight) were determined as described previously.42
Immunoblot
Liver and cell lysates were prepared and immunoblots performed as described in the supplemental data using rabbit anti-Smad1 (1:1000; 9743S; Cell Signaling), rabbit anti-Smad5 (1:1000; ab40771; Abcam), rabbit anti–phosphorylated Smad5 (pSmad5; 1:500; ab92698; Abcam [hereafter called pSmad1/5 antibody because of crossreactivity with pSmad1]), or mouse anti-actin (1:20 000; MAB1501; Millipore) antibodies. Antibodies had been verified previously43 or were verified as shown in supplemental Figure 2. Chemiluminescence quantitation of scanned films was performed using ImageJ 1.46v.
Statistics
All data are shown as mean ± standard error of the mean. Means were compared by Student unpaired t test, paired t test, or 1-way analysis of variance with Dunnett’s post hoc test using Prism 7 (GraphPad). P < .05 was considered significant.
Results
SMAD5 has a dominant role and SMAD1 a contributory role in stimulating hepcidin in Hep3B cells
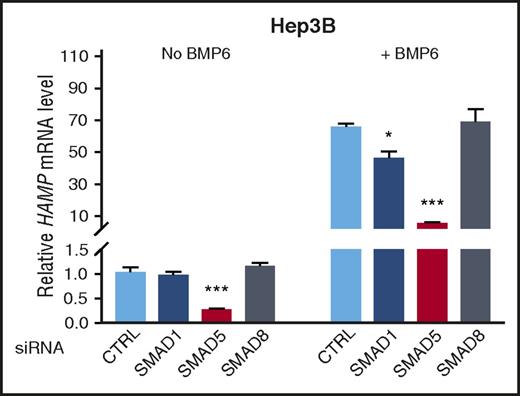
To define the relative contribution of BMP R-SMADs in hepcidin transcription, we first used siRNA to knockdown SMAD1, SMAD5, or SMAD8 and measured HAMP mRNA levels in human hepatoma Hep3B cells. Knockdown efficiency and specificity for each siRNA were verified (supplemental Figure 3). Under basal conditions, SMAD5 siRNA, but not SMAD1 or SMAD8 siRNA, inhibited HAMP mRNA expression (Figure 1 left). SMAD5 knockdown also robustly inhibited BMP6 stimulation of HAMP mRNA (∼90%), whereas SMAD1 knockdown had a modest inhibitory effect (∼30%), and SMAD8 knockdown had no significant effect (Figure 1 right). Given the dominant role of SMAD5 and, to a lesser extent, SMAD1 in regulating hepcidin expression in vitro, we therefore generated mice with a conditional knockout of Smad5, Smad1, or the combination of Smad1 and Smad5 in hepatocytes to determine their relative contributions to hepcidin expression in vivo.
SMAD5 has a dominant role and SMAD1 a contributory role in the regulation of hepcidin in Hep3B cells. Hep3B cells were transfected with negative control (CTRL), SMAD1, SMAD5, or SMAD8 siRNA (20 nM), serum starved with 1% fetal bovine serum overnight, and incubated in the absence or presence of 5 ng/ml of BMP6 for 6 hours. Relative HAMP mRNA levels were determined using quantitative reverse-transcriptase PCR. Transcripts were normalized to an internal control RPL19, and the average of CTRL without BMP6 stimulation was set to 1. Values represent mean ± standard error of the mean. *P < .05; ***P < .001 relative to the respective CTRL by 1-way analysis of variance with Dunnett’s post hoc test (n = 3-4 per group).
SMAD5 has a dominant role and SMAD1 a contributory role in the regulation of hepcidin in Hep3B cells. Hep3B cells were transfected with negative control (CTRL), SMAD1, SMAD5, or SMAD8 siRNA (20 nM), serum starved with 1% fetal bovine serum overnight, and incubated in the absence or presence of 5 ng/ml of BMP6 for 6 hours. Relative HAMP mRNA levels were determined using quantitative reverse-transcriptase PCR. Transcripts were normalized to an internal control RPL19, and the average of CTRL without BMP6 stimulation was set to 1. Values represent mean ± standard error of the mean. *P < .05; ***P < .001 relative to the respective CTRL by 1-way analysis of variance with Dunnett’s post hoc test (n = 3-4 per group).
Validation of mice with a hepatocyte-specific inactivation of Smad5 and/or Smad1
Smad1fl/fl;Smad5fl/fl mice26 were crossed with mice expressing a hepatocyte-specific Cre transgene40 to generate mice with hepatocyte-specific inactivation of Smad5 (Smad5fl/fl;Cre+), Smad1 (Smad1fl/fl;Cre+), or both (Smad1fl/fl;Smad5fl/fl;Cre+). All mice were compared with littermate Cre− controls. We also generated hepatocyte-specific Smad5 knockout mice with 1 functional allele of Smad1 (Smad1fl/wt;Smad5fl/fl;Cre+) and hepatocyte-specific Smad1 knockout mice with 1 functional allele of Smad5 (Smad1fl/fl;Smad5fl/wt;Cre+) and their respective Cre− littermate controls to study the impact of ablating 3 of 4 Smad1/5 alleles.
In the Cre+ mice, excision of the LoxP-flanked region was confirmed by PCR of genomic DNA from total liver (which includes both hepatocytes and nonparenchymal cells; Figure 2A). Quantitative reverse-transcriptase PCR analysis demonstrated that Smad5 and Smad1 mRNA levels were 77% lower in total liver and >95% lower in isolated hepatocytes of double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with littermate controls (Smad1fl/fl;Smad5fl/fl;Cre−; Figure 2B-C). Similar reductions in Smad5 and Smad1 mRNA were seen in single-knockout Smad5fl/fl;Cre+ and Smad1fl/fl;Cre+ mice compared with respective Cre− littermate controls. No compensatory increases were seen in Smad1 or Smad5 mRNA in Smad5fl/fl;Cre+ or Smad1fl/fl;Cre+ mice, respectively. Smad8 mRNA was unchanged in single-knockout mice but was reduced in double-knockout mice, consistent with previous findings that Smad8 expression is positively regulated by Bmp signaling,44 which we confirmed in Hep3B cells (supplemental Figure 4). Western blot analysis of whole-liver lysate detected an immunoreactive band for both Smad5 and Smad1 at ∼55 kDa in Smad1fl/fl;Smad5fl/fl;Cre− mice but not in Smad1fl/fl;Smad5fl/fl;Cre+ mice (Figure 2D), thus confirming the loss of hepatic Smad5 and Smad1 protein expression. Liver lysates of Smad5fl/fl;Cre+ and Smad1fl/fl;Cre+ mice also showed the loss of Smad5 and Smad1 protein expression, respectively, without compensatory increases in the remaining Smad protein (Figure 2D). A similar band pattern was detected for both Smad5 and Smad1 in primary hepatocytes isolated from these animals (data not shown).
Confirmation of hepatocyte Smad1 and/or Smad5 ablation in conditional knockout mice. (A) Schematic depictions of loxP-flanked (floxed) Smad1 or Smad5 allele and the allele after Cre recombinase-mediated excision. F and R indicate forward and reverse primers used for PCR genotyping (left). PCR analysis of genomic DNA extracted from total liver (containing both hepatocytes and nonparenchymal cells) of double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice and littermate Cre− controls at 8 weeks of age (right). (B-C) Relative Smad5 (B) and Smad1 (C) mRNA levels in the total liver (n = 7-9 per group; 8 weeks of age) and isolated hepatocytes (n = 3-4 per group; 6 weeks of age) of Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, and Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with their respective littermate Cre− controls. Transcripts were normalized to Rpl19, and the average of the respective Cre− control mice was set to 1. Values represent mean ± standard error of the mean. ***P < .001 relative to the respective Cre− controls by Student t test. (D) Western blot analysis of Smad5 and Smad1 in the livers of Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, and Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with their respective littermate Cre− controls at 8 weeks of age. Actin is used as a loading control.
Confirmation of hepatocyte Smad1 and/or Smad5 ablation in conditional knockout mice. (A) Schematic depictions of loxP-flanked (floxed) Smad1 or Smad5 allele and the allele after Cre recombinase-mediated excision. F and R indicate forward and reverse primers used for PCR genotyping (left). PCR analysis of genomic DNA extracted from total liver (containing both hepatocytes and nonparenchymal cells) of double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice and littermate Cre− controls at 8 weeks of age (right). (B-C) Relative Smad5 (B) and Smad1 (C) mRNA levels in the total liver (n = 7-9 per group; 8 weeks of age) and isolated hepatocytes (n = 3-4 per group; 6 weeks of age) of Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, and Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with their respective littermate Cre− controls. Transcripts were normalized to Rpl19, and the average of the respective Cre− control mice was set to 1. Values represent mean ± standard error of the mean. ***P < .001 relative to the respective Cre− controls by Student t test. (D) Western blot analysis of Smad5 and Smad1 in the livers of Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, and Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with their respective littermate Cre− controls at 8 weeks of age. Actin is used as a loading control.
Hepatocyte ablation of both Smad5 and Smad1 is required for developing massive iron overload in mice
To examine the relative contributions of Smad5 and Smad1 in hepcidin regulation and iron homeostasis in vivo, we first examined iron status parameters in hepatocyte-specific single-knockout Smad5fl/fl;Cre+ and Smad1fl/fl;Cre+ mice at 8 weeks of age. Serum transferrin saturation was not affected in Smad5fl/fl;Cre+ or Smad1fl/fl;Cre+ mice of either sex (Figure 3A). Liver nonheme iron concentrations did not differ in Smad5fl/fl;Cre+ or Smad1fl/fl;Cre+ male mice; however, we observed a subtle increase in liver iron levels in Smad1fl/fl;Cre+, but not in Smad5fl/fl;Cre+, female mice (Figure 3B). Next, we examined iron parameters in mice with 1 remaining functional allele of Smad1 (Smad1fl/wt;Smad5fl/fl;Cre+) or Smad5 (Smad1fl/fl;Smad5fl/wt;Cre+) in hepatocytes. We found that Smad1fl/wt;Smad5fl/fl;Cre+ female mice and Smad1fl/fl;Smad5fl/wt;Cre+ male and female mice displayed mildly elevated levels of transferrin saturation and liver iron at 8 weeks. Finally, we examined hepatocyte-specific double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice. Both male and female Smad1fl/fl;Smad5fl/fl;Cre+ mice had ∼90% transferrin saturation and developed massive liver iron overload (Figure 3C-D) at 8 weeks of age.
Hepatocyte Smad1 or Smad5 single-knockout mice exhibit minimal to no iron loading, whereas knockout of 3 or 4 Smad1/5 alleles causes progressive serum and liver iron overload. (A-B) Serum transferrin saturation (Tf sat; A) and hepatic nonheme iron concentrations (B) of Smad5fl/fl;Cre+ and Smad1fl/fl;Cre+ mice (n = 5-7 per group) compared with their respective littermate Cre− controls. (C-D) Serum transferrin saturation (C) and hepatic nonheme iron concentrations (D) of Smad1fl/wt;Smad5fl/fl;Cre+, Smad1fl/fl;Smad5fl/wt;Cre+, and double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with their respective littermate Cre− controls (n = 6-12 per group). Values represent mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 relative to the respective Cre− controls by Student t test.
Hepatocyte Smad1 or Smad5 single-knockout mice exhibit minimal to no iron loading, whereas knockout of 3 or 4 Smad1/5 alleles causes progressive serum and liver iron overload. (A-B) Serum transferrin saturation (Tf sat; A) and hepatic nonheme iron concentrations (B) of Smad5fl/fl;Cre+ and Smad1fl/fl;Cre+ mice (n = 5-7 per group) compared with their respective littermate Cre− controls. (C-D) Serum transferrin saturation (C) and hepatic nonheme iron concentrations (D) of Smad1fl/wt;Smad5fl/fl;Cre+, Smad1fl/fl;Smad5fl/wt;Cre+, and double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice compared with their respective littermate Cre− controls (n = 6-12 per group). Values represent mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 relative to the respective Cre− controls by Student t test.
Next, we measured hepatic Hamp mRNA expression in mice of each genotype. Interestingly, only double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice displayed significantly reduced Hamp mRNA expression compared with littermate Cre− mice at 8 weeks of age (Figure 4A-B). Notably, hepcidin expression is induced by iron. Hamp mRNA levels may therefore be inappropriately low relative to iron levels in Smad1fl/wt;Smad5fl/fl;Cre+ female mice and Smad1fl/fl;Smad5fl/wt;Cre+ mice that exhibit some degree of iron loading. We therefore examined Hamp mRNA levels at an earlier age before mice were exposed to the high iron content of the standard rodent diet. Although mice are typically weaned at 3 weeks of age, they can start nibbling solid food as early as 2 weeks old. Therefore, we harvested mice at 12 days to minimize the effect of dietary iron. At 12 days, Hamp mRNA levels were significantly reduced in all Cre+ mice (Figure 4C-D), when liver iron content was not yet increased (supplemental Figure 5).
Liver Hamp mRNA is reduced at 12 days of age in mice lacking 2 to 4 Smad1/5 alleles but is reduced at 8 weeks of age only in double-knockout mice. Relative expression of Hamp was measured in Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, Smad1fl/wt;Smad5fl/fl;Cre+, Smad1fl/fl;Smad5fl/wt;Cre+, Smad1fl/fl;Smad5fl/fl;Cre+, and littermate control Cre− mouse livers at 8 weeks (A-B; n = 5-11 per group) or 12 days (C-D; n = 3-6 per group) of age. Transcript levels were normalized to Rpl19, and the average of the respective littermate Cre− control mice was set to 1. Values represent mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 relative to the respective Cre− controls by Student t test.
Liver Hamp mRNA is reduced at 12 days of age in mice lacking 2 to 4 Smad1/5 alleles but is reduced at 8 weeks of age only in double-knockout mice. Relative expression of Hamp was measured in Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, Smad1fl/wt;Smad5fl/fl;Cre+, Smad1fl/fl;Smad5fl/wt;Cre+, Smad1fl/fl;Smad5fl/fl;Cre+, and littermate control Cre− mouse livers at 8 weeks (A-B; n = 5-11 per group) or 12 days (C-D; n = 3-6 per group) of age. Transcript levels were normalized to Rpl19, and the average of the respective littermate Cre− control mice was set to 1. Values represent mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 relative to the respective Cre− controls by Student t test.
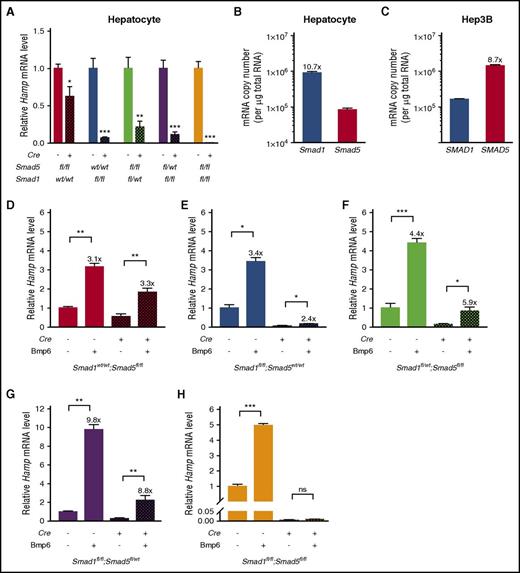
We then tested the ability of primary hepatocytes from mice of each genotype to respond to Bmp6 stimulation. Consistent with total liver Hamp mRNA levels in 12-day-old mice, baseline Hamp mRNA levels were significantly reduced in primary hepatocytes of all Cre+ genotypes (Figure 5A). In contrast to the findings in human Hep3B cells where SMAD5 had a dominant role (Figure 1), Smad1 seemed to have a greater role in baseline Hamp mRNA expression in mouse primary hepatocytes, because primary hepatocytes from Smad1fl/fl;Cre+ and Smad1fl/fl;Smad5fl/wt;Cre+ mice had greater reductions in baseline Hamp mRNA relative to littermate Cre− mice compared with Smad5fl/fl;Cre+ and Smad1fl/wt;Smad5fl/fl;Cre+ mice, respectively. The higher expression of Smad1 relative to Smad5 in mouse hepatocytes compared with the lower expression of SMAD1 relative to SMAD5 in human Hep3B cells (Figure 5B-C) may explain the apparent differences between the functional role of R-SMADs in hepcidin regulation in these systems.
Hepcidin is induced by Bmp6 in primary hepatocytes from mice lacking 2 or 3 Smad1/5 alleles but not in double-knockout mice. (A,D-H) Primary hepatocytes were isolated from 6-week-old male Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, Smad1fl/wt;Smad5fl/fl;Cre+, Smad1fl/fl;Smad5fl/wt;Cre+, Smad1fl/fl;Smad5fl/fl;Cre+, and littermate Cre− control mice using 2-step collagenase digestion and cultured in collagen-coated plates. Cells were serum starved with 1% fetal bovine serum overnight before Bmp6 stimulation for 6 hours. Hepatocyte Hamp mRNA levels were determined at baseline (A; n = 4-5 per group) and after stimulation with Bmp6 (D-H; n = 3-5 per group) for each genotype. Transcript levels were normalized to Rpl19, and the average of the respective Cre− control mice without Bmp6 stimulation was set to 1. Values represent mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 relative to the respective Cre− controls (A) or unstimulated cells of the same genotype (D-H) by Student t test. Fold induction of Hamp mRNA in Bmp6-stimulated versus -nonstimulated cells for each genotype is indicated. (B-C) Smad1 and Smad5 mRNA copy numbers were determined using Taqman primers with probes targeting all known variants and quantitative reverse-transcriptase PCR in mouse primary hepatocytes (B) and human Hep3B cells (C; n = 4 per group). ns, not significant.
Hepcidin is induced by Bmp6 in primary hepatocytes from mice lacking 2 or 3 Smad1/5 alleles but not in double-knockout mice. (A,D-H) Primary hepatocytes were isolated from 6-week-old male Smad5fl/fl;Cre+, Smad1fl/fl;Cre+, Smad1fl/wt;Smad5fl/fl;Cre+, Smad1fl/fl;Smad5fl/wt;Cre+, Smad1fl/fl;Smad5fl/fl;Cre+, and littermate Cre− control mice using 2-step collagenase digestion and cultured in collagen-coated plates. Cells were serum starved with 1% fetal bovine serum overnight before Bmp6 stimulation for 6 hours. Hepatocyte Hamp mRNA levels were determined at baseline (A; n = 4-5 per group) and after stimulation with Bmp6 (D-H; n = 3-5 per group) for each genotype. Transcript levels were normalized to Rpl19, and the average of the respective Cre− control mice without Bmp6 stimulation was set to 1. Values represent mean ± standard error of the mean. *P < .05; **P < .01; ***P < .001 relative to the respective Cre− controls (A) or unstimulated cells of the same genotype (D-H) by Student t test. Fold induction of Hamp mRNA in Bmp6-stimulated versus -nonstimulated cells for each genotype is indicated. (B-C) Smad1 and Smad5 mRNA copy numbers were determined using Taqman primers with probes targeting all known variants and quantitative reverse-transcriptase PCR in mouse primary hepatocytes (B) and human Hep3B cells (C; n = 4 per group). ns, not significant.
Although basal levels were reduced, Hamp mRNA was induced by Bmp6 in all single-knockout and homozygous/heterozygous Smad1fl/fl;Smad5fl/wt;Cre+ and Smad1fl/wt;Smad5fl/fl;Cre+ strains (Figure 5D-G). Hamp mRNA levels were lower in Bmp6-stimulated Cre+ hepatocytes compared with Bmp6-stimulated Cre− hepatocytes; however, the fold increases relative to unstimulated cells of the same genotype were generally similar. This preserved inducibility of Hamp may help account for the fact that Hamp mRNA levels in these mice seemed similar to littermate Cre− mice at 8 weeks of age after exposure to dietary iron, when Bmp signaling is induced. In contrast, Hamp mRNA levels failed to be induced by Bmp6 in double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ primary hepatocytes (Figure 5H).
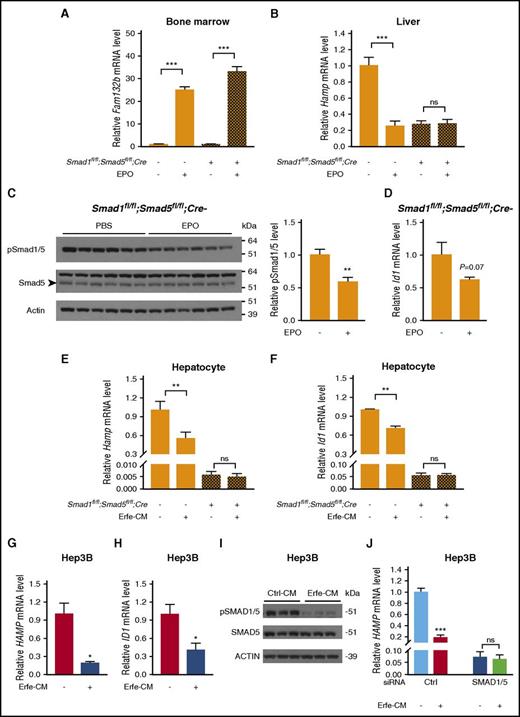
Smad1/5 is required for EPO and erythroferrone suppression of hepcidin in mice
To determine if erythropoietic suppression of hepcidin requires the BMP-SMAD pathway, we tested the effects of EPO injection in Smad1fl/fl;Smad5fl/fl;Cre+ mice. Consistent with previous findings,14,45 EPO similarly induced erythroferrone (Fam132b) mRNA in the bone marrow of Smad1fl/fl;Smad5fl/fl;Cre+ mice and littermate controls (Figure 6A). However, whereas EPO robustly reduced liver Hamp mRNA expression in Smad1fl/fl;Smad5fl/fl;Cre− mice, it did not suppress Hamp in Smad1fl/fl;Smad5fl/fl;Cre+ mice (Figure 6B). Similar results were seen for serum hepcidin (supplemental Figure 6). This suggests that EPO requires an intact SMAD1/5 signaling pathway to suppress hepcidin expression. EPO suppression of hepcidin was preserved in single-knockout Smad5fl/fl;Cre+and Smad1fl/fl;Cre+ mice, suggesting that Smad1 and Smad5 function redundantly in EPO-mediated hepcidin suppression (supplemental Figure 7). Notably, liver Smad1/5 phosphorylation was significantly decreased in EPO-treated Smad1fl/fl;Smad5fl/fl;Cre− mice compared with phosphate-buffered saline–treated mice (Figure 6C), similar to results in 1 prior report.45 A trend toward lower mRNA expression of the Bmp-Smad1/5 target Id1 was also observed in response to EPO treatment, but it did not reach statistical significance (P = .07; Figure 6D). These data raise the possibility that EPO may have a functional role in suppressing Smad pathway activity in the liver.
Smad1 and Smad5 are required for EPO and erythroferrone suppression of hepcidin in mice.Smad1fl/fl;Smad5fl/fl;Cre+ and littermate Cre− mice at 6 weeks of age were injected with phosphate-buffered saline (PBS) or EPO (200 U per mouse), and tissues were harvested after 15 hours to determine bone marrow Fam132b mRNA expression (A), liver Hamp mRNA (B), liver phosphorylated Smad1/5 protein (C), and liver Id1 mRNA levels (D). Primary hepatocytes isolated from 6-week-old male Smad1fl/fl;Smad5fl/fl;Cre+ and littermate Cre− mice (E-F) and Hep3B cells (G-I) were treated with conditioned medium containing 50% (volume-to-volume ratio) cell supernatant from control HEK293T cells (Ctrl-CM) or Erfe-CM for 15 hours (E-F) or 6 hours (G-I), and the relative mRNA levels of Hamp (E,G), Id1 (F,H), and pSMAD1/5 protein (I) were determined. (J) Hep3B cells were transfected with control siRNA or SMAD1 and SMAD5 siRNA for 48 hours before 6-hour Ctrl-CM or Erfe-CM treatment, and relative HAMP mRNA levels were determined. Transcript levels measured by quantitative reverse-transcriptase PCR were normalized to Rpl19, Smad1/5 phosphorylation levels determined by immunoblot were normalized to total Smad5, and the average of PBS-treated Cre− control mice or Ctrl-CM–treated cells was set to 1. Representative immunoblots are shown. Values represent mean ± standard error of the mean (n = 4-5 mice per group in panel A; n = 10 mice per group in panels B-D; n = 4 per group in panels E-F; n = 3 per group in panels G-J). *P < .05; **P < .01; ***P < .001 relative to PBS-treated mice or Ctrl-CM–treated cells of the same genotype by Student t test. ns, not significant.
Smad1 and Smad5 are required for EPO and erythroferrone suppression of hepcidin in mice.Smad1fl/fl;Smad5fl/fl;Cre+ and littermate Cre− mice at 6 weeks of age were injected with phosphate-buffered saline (PBS) or EPO (200 U per mouse), and tissues were harvested after 15 hours to determine bone marrow Fam132b mRNA expression (A), liver Hamp mRNA (B), liver phosphorylated Smad1/5 protein (C), and liver Id1 mRNA levels (D). Primary hepatocytes isolated from 6-week-old male Smad1fl/fl;Smad5fl/fl;Cre+ and littermate Cre− mice (E-F) and Hep3B cells (G-I) were treated with conditioned medium containing 50% (volume-to-volume ratio) cell supernatant from control HEK293T cells (Ctrl-CM) or Erfe-CM for 15 hours (E-F) or 6 hours (G-I), and the relative mRNA levels of Hamp (E,G), Id1 (F,H), and pSMAD1/5 protein (I) were determined. (J) Hep3B cells were transfected with control siRNA or SMAD1 and SMAD5 siRNA for 48 hours before 6-hour Ctrl-CM or Erfe-CM treatment, and relative HAMP mRNA levels were determined. Transcript levels measured by quantitative reverse-transcriptase PCR were normalized to Rpl19, Smad1/5 phosphorylation levels determined by immunoblot were normalized to total Smad5, and the average of PBS-treated Cre− control mice or Ctrl-CM–treated cells was set to 1. Representative immunoblots are shown. Values represent mean ± standard error of the mean (n = 4-5 mice per group in panel A; n = 10 mice per group in panels B-D; n = 4 per group in panels E-F; n = 3 per group in panels G-J). *P < .05; **P < .01; ***P < .001 relative to PBS-treated mice or Ctrl-CM–treated cells of the same genotype by Student t test. ns, not significant.
Because EPO acts indirectly to suppress hepcidin and could involve multiple mechanisms, we isolated primary hepatocytes from Smad1fl/fl;Smad5fl/fl;Cre+ mice and littermate controls to examine more specifically whether erythroferrone suppression of hepcidin requires Smad1/5. Whereas treatment with conditioned medium from HEK293T cells overexpressing erythroferrone (Erfe-CM)14 or transfection with Fam132b cDNA significantly decreased Hamp and Id1 mRNA levels in primary hepatocytes from Smad1fl/fl;Smad5fl/fl;Cre− mice, neither Hamp nor Id1 mRNA was reduced in hepatocytes from Smad1fl/fl;Smad5fl/fl;Cre+ mice (Figure 6E-F; supplemental Figure 8A-C). To examine a more direct effect for erythroferrone on Smad1/5 phosphorylation, we treated Hep3B cells with Erfe-CM for 6 hours. Similar to mouse primary hepatocytes, Hep3B cells treated with Erfe-CM had reduced HAMP and ID1 mRNA (Figure 6G-H), and notably, pSMAD1/5 was reduced ∼75% (Figure 6I). Fam132b transfection likewise inhibited pSMAD1/5 protein, HAMP, and ID1 mRNA in Hep3B cells (supplemental Figure 8D-F). Consistent with the primary hepatocyte data, siRNA-mediated knockdown of SMAD1 and SMAD5 blocked the ability of Erfe-CM to suppress HAMP mRNA in Hep3B cells (Figure 6J).
To rule out the possibility that basal hepcidin expression in Smad1fl/fl;Smad5fl/fl;Cre+ mice was not suppressible by EPO because it had already reached its nadir, we investigated whether hepcidin was suppressed by dietary iron deficiency in these mice. Liver Hamp mRNA, serum hepcidin, and liver iron levels were significantly lower in Smad1fl/fl;Smad5fl/fl;Cre+ mice fed a low-iron diet for 3 weeks after weaning compared with Smad1fl/fl;Smad5fl/fl;Cre+ mice fed a standard diet (Figure 7A-B left; supplemental Figure 9). Thus, hepcidin is further suppressible in Smad1fl/fl;Smad5fl/fl;Cre+ mice in some contexts. Interestingly, treatment with neutralizing BMP2/4 and BMP6 antibodies completely blocked the residual ability of iron in the standard diet to increase Hamp mRNA in Smad1fl/fl;Smad5fl/fl;Cre+ mice, leading to more liver iron loading (Figure 7A-B right). This suggests that residual hepcidin induction by iron in the Smad1fl/fl;Smad5fl/fl;Cre+ mice is dependent on residual Bmp signaling.
Hamp mRNA levels are suppressed in double-knockout mice treated with a low-iron diet. (A-B left) Smad1fl/fl;Smad5fl/fl;Cre+ mice and littermate Cre− controls were fed a standard or low-iron diet (FeD) for 3 weeks upon weaning. (A-B right) Smad1fl/fl;Smad5fl/fl;Cre+ mice were fed a low-iron diet for 2 weeks upon weaning and switched to standard diet for 1 week, coupled with 4 intraperitoneal injections of phosphate-buffered saline (PBS) or a mixture of neutralizing BMP2/4 (10 mg/kg of body weight) and BMP6 (5 mg/kg of body weight) antibodies in PBS administered every other day. Livers were harvested to measure Hamp mRNA levels (A) and nonheme iron concentrations (B; n = 3-8 per group). Values represent mean ± standard error of the mean. *P < .05; **P < .01 relative to mice on a standard diet of the same genotype (left) or relative to PBS-treated mice (right) by Student t test.
Hamp mRNA levels are suppressed in double-knockout mice treated with a low-iron diet. (A-B left) Smad1fl/fl;Smad5fl/fl;Cre+ mice and littermate Cre− controls were fed a standard or low-iron diet (FeD) for 3 weeks upon weaning. (A-B right) Smad1fl/fl;Smad5fl/fl;Cre+ mice were fed a low-iron diet for 2 weeks upon weaning and switched to standard diet for 1 week, coupled with 4 intraperitoneal injections of phosphate-buffered saline (PBS) or a mixture of neutralizing BMP2/4 (10 mg/kg of body weight) and BMP6 (5 mg/kg of body weight) antibodies in PBS administered every other day. Livers were harvested to measure Hamp mRNA levels (A) and nonheme iron concentrations (B; n = 3-8 per group). Values represent mean ± standard error of the mean. *P < .05; **P < .01 relative to mice on a standard diet of the same genotype (left) or relative to PBS-treated mice (right) by Student t test.
Discussion
BMP-SMAD signaling is a central pathway in the regulation of hepcidin transcription.11,22 Although the contributions of specific BMP type I and type II receptors have been described,20,21 the role of individual R-SMADs remains to be elucidated. Our finding that hepatocyte-specific double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice developed massive liver iron overload but single-knockout Smad5fl/fl;Cre+ or Smad1fl/fl;Cre+ mice had minimal to no iron loading at 8 weeks of age suggest that hepatocyte Smad1 and Smad5 have overlapping functions and work collaboratively to govern Hamp expression in the liver. The intermediate phenotype of Smad1fl/wt;Smad5fl/fl;Cre+ and Smad1fl/fl;Smad5fl/wt;Cre+ mice with 1 remaining functional Smad1 or Smad5 allele suggests a gene dosage effect. Although Smad1 and Smad5 have nonoverlapping functions in some contexts,28 these observations are consistent with many other studies where Smad1 and Smad5 were shown to have dose-dependent yet redundant functions, for example, in mouse embryo during early development,25 in gonadal somatic cells for reproduction,26 and in chondrocyte differentiation for bone formation.27
The single-knockout Smad5fl/fl;Cre+ or Smad1fl/fl;Cre+ mice and mice with 1 remaining functional allele of Smad1 or Smad5 in hepatocytes (Smad1fl/wt;Smad5fl/fl;Cre+ and Smad1fl/fl;Smad5fl/wt;Cre+ mice, respectively) provide models to study the effects of more subtle defects in the BMP-SMAD signaling pathway. Liver Hamp expression was significantly reduced in all these models at 12 days of age as well as in isolated primary hepatocyte from 6-week-old animals, providing evidence that Smad1 and Smad5 have a role in maintaining basal Hamp mRNA levels. We attribute the lack of significant iron phenotype at 8 weeks of age in mice with 2 of 4 functional Smad1 and Smad5 alleles to the preserved inducibility of Hamp under Bmp6 and dietary iron exposure. Mice with 1 of 4 functional Smad1 and Smad5 alleles required higher iron levels to induce Hamp levels equivalent to those in Cre− littermate controls, thereby resulting in a mild iron overload phenotype. However, even the presence of 1 Smad1 or Smad5 allele was enough for hepcidin regulation to remain largely intact.
One apparent discrepancy between our in vitro and in vivo studies was that SMAD5 had a dominant role in HAMP regulation by BMP6 in Hep3B cells, whereas Smad1 had a slightly more prominent role in mice, because Smad1fl/fl;Smad5fl/wt;Cre+ mice seemed to have lower hepcidin levels and a stronger iron phenotype than Smad1fl/wt;Smad5fl/fl;Cre+ mice, at least in males. Notably, quantitative analysis revealed higher levels of SMAD5 versus SMAD1 in Hep3B cells compared with lower levels of Smad5 versus Smad1 in mouse primary hepatocytes. These data suggest that both Smad1 and Smad5 can regulate Hamp, and we hypothesize that the difference between these models results from the relative expression of Smad1 compared with Smad5. Given the differences between our models, we cannot exclude the possibility that Smad8 may also have a role in hepcidin regulation and iron homeostasis in vivo, even though SMAD8 knockdown did not inhibit HAMP expression in Hep3B cells. Previous work has indicated that Smad8 has insignificant functions in embryo development and cartilage formation as a result of the redundancy with Smad1 and Smad5.25,27 However, Smad8 expression is increased by the activation of Bmp signaling and, at least in vitro, can function to suppress Bmp signaling by forming a heterodimer with Smad1 and Smad5 to inhibit their effects.44,46 Whether Smad8 works to accelerate or antagonize Bmp signaling in the liver is currently under investigation.
The Smad1fl/fl;Smad5fl/fl;Cre+ mice provide a model to study how the BMP R-SMAD pathway interacts with other hepcidin regulators for which the mechanism of action is less well understood. For example, we recently used these mice to demonstrate that hepcidin induction by endoplasmic reticulum stress requires Smad1/5 signaling.47 Here, we explored the role of Smad1/5 in hepcidin suppression by erythropoietic drive. Several secreted proteins have been proposed to function as erythroid regulators of hepcidin, including growth differentiation factor 15,48 twisted gastrulation BMP signaling modulator 1,49 and erythroferrone.14 On the basis of recent studies, growth differentiation factor 15 and twisted gastrulation BMP signaling modulator 1 do not seem to be physiological suppressors of hepcidin.14,50-52 In contrast, erythroferrone seems essential, because Fam132b-knockout animals did not suppress hepcidin in response to acute erythropoiesis and had delayed recovery of hemoglobin after hemorrhage.14 Here, we found that EPO and erythroferrone did not further suppress hepcidin expression in hepatocytes lacking both Smad1 and Smad5, suggesting that Smad1/5 are required for EPO- and erythroferrone-mediated suppression of hepcidin. Previous work has reported that EPO suppression of hepcidin is not impaired in mice deficient for Tfr2, Hfe2, or Bmp6,14,45,53 nor in mice fed an iron-deficient diet for 3 weeks, all of which lead to suppressed Smad signaling in the liver. One possibility is that EPO and erythroferrone act further downstream in the Smad signaling cascade, or the residual Smad expression in these models may be enough to permit the hepcidin suppressive effects. Not only does a loss of Smad1/5 block EPO and erythroferrone suppression of hepcidin, but EPO effects are also blunted when the Smad1/5 pathway is highly induced, for example, in Tmprss6-knockout mice and mice fed a high-iron diet.45 Similarly, in the context of thalassemia, where high erythropoietin/erythroferrone levels and iron overload coexist, Hamp mRNA was more suppressed in Th3/+ mice maintained on a low-iron diet to achieve normal liver iron levels compared with mice on an iron-sufficient diet with iron overload.54 Thus, Smad1/5 signaling must be appropriately regulated for the maximal hepcidin suppressive effects of EPO and erythroferrone.
Interestingly, EPO injection in mice led to reduced liver pSmad1/5 expression and a strong trend toward lower expression of the Smad1/5 target transcript Id1, suggesting that 1 mechanism by which erythroferrone suppresses hepcidin may be by inhibiting Smad1/5 signaling. Although the initial report describing erythroferrone failed to detect reduced liver pSmad5 in mice treated with EPO or phlebotomy, decreased Id1 expression was seen after phlebotomy.14 Moreover, a recent study reported reduced liver pSmad5 and Id1 after EPO injection in wild-type mice.45 Additionally, several studies have described a failure to appropriately induce pSmad1/5 by iron overload in the context of ineffective erythropoiesis in thalassemia.52,55 However, a direct effect of erythroferrone on Smad1/5 signaling has not previously been examined. Here, we demonstrated that treatment with erythroferrone conditioned medium or transfection with Fam132b cDNA decreased SMAD1/5 phosphorylation and ID1 expression in Hep3B cells in 6 hours, suggesting that erythroferrone does have a functional role in suppressing SMAD1/5 signaling in hepatocytes. Although it has been proposed that Tmprss6 mediates the liver pSmad1/5 reduction in response to EPO required for hepcidin suppression,45 a recent correspondence reported that erythroferrone still suppressed Hamp and Id1 mRNA in Tmprss6−/− primary hepatocytes,56 suggesting a Tmprss6-independent effect. Future studies will be needed to understand the molecular mechanisms by which erythroferrone suppresses Smad1/5 signaling and the full details of crosstalk between these pathways.
In contrast to the effects of EPO injection, a low-iron diet further suppressed hepcidin in double-knockout Smad1fl/fl;Smad5fl/fl;Cre+ mice. Interestingly, the residual ability of iron to increase hepcidin in the Smad1fl/fl;Smad5fl/fl;Cre+ mice on a standard diet compared with a low-iron diet was blocked by neutralizing BMP2/4 and BMP6 antibodies, suggesting that residual hepcidin induction by iron in these mice is dependent on residual Bmp signaling. Because Cre-mediated recombination is not 100% efficient, and because Smad8 is still present, albeit at reduced levels, residual iron-mediated hepcidin expression in these mice may be governed by residual Smad1/5/8 signaling. Alternatively, a noncanonical pathway activated by BMPs could be involved.57 However, this residual Bmp signaling in the Smad1fl/fl;Smad5fl/fl;Cre+ mice does not seem to be sufficient to mediate hepcidin suppression by erythropoietin.
In summary, our results demonstrate that Smad1 and Smad5 have redundant, dose-dependent roles in hepcidin regulation and iron homeostasis. Moreover, Smad1 and Smad5 are required for the suppressive effects of erythropoietin and erythroferrone on hepcidin to optimize iron availability for red blood cell production in the context of anemia. The Smad1fl/fl;Smad5fl/fl;Cre+ mice provide a new tool to understand how various signals are integrated to regulate hepcidin expression and govern systemic iron homeostasis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elizabeth Robertson (University of Oxford, Oxford, United Kingdom) for kindly providing the Smad1 floxed mice and Tomas Ganz and Elizabeta Nemeth (University of California Los Angeles, Los Angeles, CA) for graciously providing HEK293T cells overexpressing erythroferrone and the conditioned media.
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants RO1-DK087727 (J.L.B.) and T32 DK007540 (K.B.Z.-B. and A.B.C.) and a Howard Goodman Fellowship Award from the Massachusetts General Hospital (J.L.B.).
Authorship
Contribution: C.-Y.W. performed experiments, interpreted data, and wrote the paper; A.B.C. initiated the generation of the hepatocyte-specific Smad1/5 double-knockout mice and performed experiments; S.C. and S.O. assisted in mouse studies; K.B.Z.-B. performed knockdown studies; L.U. and A.Z. provided key reagents; and J.L.B. conceived and oversaw the study, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: J.L.B. has ownership interest in Ferrumax Pharmaceuticals, which has licensed technology from the Massachusetts General Hospital based on work cited here and in prior publications. The remaining authors declare no competing financial interests.
Correspondence: Jodie L. Babitt, Massachusetts General Hospital, 185 Cambridge St, CPZN-8208, Boston, MA 02114; e-mail: babitt.jodie@mgh.harvard.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal