Abstract
Molecular diagnostics has generated substantial dividends in dissecting the genetic basis of myeloid neoplasms with eosinophilia. The family of diseases generated by dysregulated fusion tyrosine kinase (TK) genes is recognized by the World Health Organization (WHO) category, “Myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, or FGFR1, or with PCM1-JAK2.” In addition to myeloproliferative neoplasms (MPN), these patients can present with myelodysplastic syndrome/MPN, as well as de novo or secondary mixed-phenotype leukemias or lymphomas. Eosinophilia is a common, but not invariable, feature of these diseases. The natural history of PDGFRA- and PDGFRB-rearranged neoplasms has been dramatically altered by imatinib. In contrast, patients with FGFR1 and JAK2 fusion TK genes exhibit a more aggressive course and variable sensitivity to current TK inhibitors, and in most cases, long-term disease-free survival may only be achievable with allogeneic hematopoietic stem cell transplantation. Similar poor prognosis outcomes may be observed with rearrangements of FLT3 or ABL1 (eg, both of which commonly partner with ETV6), and further investigation is needed to validate their inclusion in the current WHO-defined group of eosinophilia-associated TK fusion-driven neoplasms. The diagnosis chronic eosinophilic leukemia, not otherwise specified (CEL, NOS) is assigned to patients with MPN with eosinophilia and nonspecific cytogenetic/molecular abnormalities and/or increased myeloblasts. Myeloid mutation panels have identified somatic variants in patients with a provisional diagnosis of hypereosinophilia of undetermined significance, reclassifying some of these cases as eosinophilia-associated neoplasms. Looking forward, one of the many challenges will be how to use the results of molecular profiling to guide prognosis and selection of actionable therapeutic targets.
Introduction
Eosinophilia is observed in a range of reactive and clonal disorders and may be associated with life-threatening organ damage as a result of infiltration of eosinophils and release of granular contents. Hypereosinophilia (HE) has been historically defined as a persistent eosinophil count of at least 1.5 × 109/L. In 2012, an international consensus group published modified criteria for peripheral blood and tissue HE and generated nomenclature for subtypes of HE.1 These include primary (clonal/neoplastic, HEN) HE, secondary HE (reactive, HER), hereditary HE (familial, HEFA), and HE of undetermined significance (HEUS), which was introduced as a term in lieu of the phrase “idiopathic hypereosinophilia” (Table 1). Idiopathic HES, a term first coined by Hardy and Anderson in 19682 and defined by Chusid and colleagues in 1975,3 denotes the presence of HE-associated organ damage without an identifiable underlying cause. The international consensus group definitions for HE and its subtypes are also provided in Table 1.
International consensus group definitions for HE and HES
| Proposed term . | Proposed abbreviation . | Pathogenesis/definition . |
|---|---|---|
| Hypereosinophilia | HE | >1.5 × 109/L eosinophils in the blood on 2 examinations (interval >1 mo*) and/or tissue HE defined by the following:† Percentage of eosinophils in bone marrow (BM) section exceeds 20% of all nucleated cells; and/or Pathologist is of the opinion that tissue infiltration by eosinophils is extensive; and/or Marked deposition of eosinophil granule proteins is found (in the absence or presence of major tissue infiltration by eosinophils). |
| Subtypes of HE | ||
| Hereditary (familial) HE | HEFA | Pathogenesis unknown; familial clustering, no signs or symptoms of hereditary immunodeficiency, and no evidence of a reactive or neoplastic condition/disorder underlying HE |
| HE of undetermined significance | HEUS | No underlying cause of HE, no family history, no evidence of a reactive or neoplastic condition/disorder underlying HE, and no end-organ damage attributable to HE |
| Primary (clonal/neoplastic) HE¶ | HEN | Underlying stem cell, myeloid, or eosinophilic neoplasm, as classified by WHO criteria; eosinophils considered neoplastic cells‡ |
| Secondary (reactive) HE¶ | HER | Underlying condition/disease in which eosinophils are considered nonclonal cells§; HE considered cytokine-driven in most cases§ |
| Hypereosinophilic syndrome | HES | Criteria for peripheral blood HE fulfilled*; and Organ damage and/or dysfunction attributable to tissue HE#; and Exclusion of other disorders or conditions as major reason for organ damage |
| Proposed term . | Proposed abbreviation . | Pathogenesis/definition . |
|---|---|---|
| Hypereosinophilia | HE | >1.5 × 109/L eosinophils in the blood on 2 examinations (interval >1 mo*) and/or tissue HE defined by the following:† Percentage of eosinophils in bone marrow (BM) section exceeds 20% of all nucleated cells; and/or Pathologist is of the opinion that tissue infiltration by eosinophils is extensive; and/or Marked deposition of eosinophil granule proteins is found (in the absence or presence of major tissue infiltration by eosinophils). |
| Subtypes of HE | ||
| Hereditary (familial) HE | HEFA | Pathogenesis unknown; familial clustering, no signs or symptoms of hereditary immunodeficiency, and no evidence of a reactive or neoplastic condition/disorder underlying HE |
| HE of undetermined significance | HEUS | No underlying cause of HE, no family history, no evidence of a reactive or neoplastic condition/disorder underlying HE, and no end-organ damage attributable to HE |
| Primary (clonal/neoplastic) HE¶ | HEN | Underlying stem cell, myeloid, or eosinophilic neoplasm, as classified by WHO criteria; eosinophils considered neoplastic cells‡ |
| Secondary (reactive) HE¶ | HER | Underlying condition/disease in which eosinophils are considered nonclonal cells§; HE considered cytokine-driven in most cases§ |
| Hypereosinophilic syndrome | HES | Criteria for peripheral blood HE fulfilled*; and Organ damage and/or dysfunction attributable to tissue HE#; and Exclusion of other disorders or conditions as major reason for organ damage |
In the case of evolving life-threatening end-organ damage, the diagnosis can be made immediately to avoid delay in therapy.
Validated quantitative criteria for tissue HE do not exist for most tissues at the present time. As a consequence, tissue HES is defined by a combination of qualitative and semiquantitative findings that will require revision as new information becomes available.
Clonality of eosinophils is often difficult to demonstrate or is not examined. However, if a myeloid or stem cell neoplasm known to present typically with clonal HE is present or atypical molecular defect is demonstrable (eg, PDGFR or FGFR mutations or BCR/ABL1), eosinophilia should be considered clonal.
HEN and HER are prediagnostic checkpoints that should guide further diagnostic evaluations but cannot serve as final diagnoses.
In a group of patients, HER might be caused/triggered by other as-yet-unknown processes because no increase in eosinophilopoietic cytokine levels can be documented.
HE-related organ damage (damage attributable to HE): organ dysfunction with marked tissue eosinophil infiltrates and/or extensive deposition of eosinophil-derived proteins (in the presence or absence of marked tissue eosinophils) and 1 or more of the following: fibrosis (lung, heart, digestive tract, skin, and others); thrombosis with or without thromboembolism; cutaneous (including mucosal) erythema, edema/angioedema, ulceration, pruritus, and eczema; and peripheral or central neuropathy with chronic or recurrent neurologic deficit. Less commonly, other organ system involvement (liver, pancreas, kidney, and other organs) and the resulting organ damage can be judged as HE-related pathology, so the clinician concludes the clinical situation resembles HES. Note that HES can manifest in 1 or more organ systems.
In the majority of cases, eosinophilia is reactive through overproduction of eosinophilopoietic cytokines such as interleukin 3 (IL-3), IL-5, or granulocyte-macrophage colony-stimulating factor in association with atopic conditions/allergies, infections, medications, autoimmune disorders, and/or rarely hematologic or solid malignancies.4 Clonal eosinophilia is most frequently associated with chronic myeloid neoplasms (eg, myeloproliferative neoplasms [MPN-eo] or myelodysplastic/myeloproliferative neoplasms [MDS/MPN-eo]), and more rarely with acute myeloid leukemia (AML) and B- or T-cell acute lymphoblastic leukemia (ALL)/lymphoma.5,6 Lymphocyte-variant HE is a reactive eosinophilia resulting from excess production of eosinophilopoietic cytokines secreted by immunophenotypically aberrant clones of T cells.7,8 If none of the aforementioned conditions is identified, a provisional diagnosis of HEUS (or HES) is rendered until a cause of eosinophilia emerges.1,3
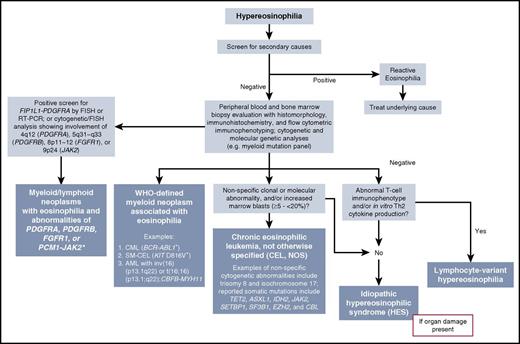
In recognition of the growing list of recurrent, genetically defined eosinophilias driven by constitutively activated tyrosine kinase (TK) fusion genes, the World Health Organization (WHO) created a new major category in 2008 termed “Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of platelet-derived growth factor receptor α (PDGFRA), platelet-derived growth factor receptor β (PDGFRB), or fibroblast growth factor receptor 1 (FGFR1).”9 This category has been revised in the WHO 2016 classification with the addition of the provisional entity of PCM1-JAK2-positive neoplasms.10 “Chronic eosinophilic leukemia, not otherwise specified” (CEL, NOS) is 1 of 7 diseases included in the WHO category of MPNs.11 A classification scheme and diagnostic algorithm for these eosinophilia-associated myeloid and/or lymphoid neoplasms within the context of the 2016 revised WHO scheme of hematolymphoid diseases are shown in Figure 1 and Table 2.
Diagnostic algorithm for HE incorporating 2016 WHO categories of eosinophilia-associated neoplasms. In patients presenting with HE, we recommend an initial work-up to rule out reactive (secondary) causes. If this work-up is negative, parallel or sequential testing for a primary (clonal) eosinophilia should be undertaken. Evaluation includes morphologic analysis of the peripheral blood and BM, immunohistochemistry (eg, CD117, tryptase, and CD25 in systemic mastocytosis), flow cytometric immunophenotyping to assess the presence of myeloid, B- and/or T-lymphocyte markers, and cytogenetic/molecular/genetic testing. As shown in the algorithm, this combination of clinicopathologic and molecular testing can help identify a specific neoplasm and the WHO category within which it resides (shown in blue-colored boxes). Idiopathic HES is a diagnosis of exclusion and requires the presence of organ damage. HES is considered a provisional diagnosis until a cause of HE is discovered. *The provisional variants ETV6-JAK2 and BCR-JAK2 are also included in this category. Figure modified from: Gotlib J, Cools J, Malone JM 3rd, et al. Blood. 2004;103(8):2879-2891.
Diagnostic algorithm for HE incorporating 2016 WHO categories of eosinophilia-associated neoplasms. In patients presenting with HE, we recommend an initial work-up to rule out reactive (secondary) causes. If this work-up is negative, parallel or sequential testing for a primary (clonal) eosinophilia should be undertaken. Evaluation includes morphologic analysis of the peripheral blood and BM, immunohistochemistry (eg, CD117, tryptase, and CD25 in systemic mastocytosis), flow cytometric immunophenotyping to assess the presence of myeloid, B- and/or T-lymphocyte markers, and cytogenetic/molecular/genetic testing. As shown in the algorithm, this combination of clinicopathologic and molecular testing can help identify a specific neoplasm and the WHO category within which it resides (shown in blue-colored boxes). Idiopathic HES is a diagnosis of exclusion and requires the presence of organ damage. HES is considered a provisional diagnosis until a cause of HE is discovered. *The provisional variants ETV6-JAK2 and BCR-JAK2 are also included in this category. Figure modified from: Gotlib J, Cools J, Malone JM 3rd, et al. Blood. 2004;103(8):2879-2891.
Revised 2016 WHO classification of eosinophilia-associated myeloid or lymphoid neoplasms and idiopathic hypereosinophilic syndrome
| Myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, or FGFR1, or with PCM1-JAK2 . |
|---|
| Diagnostic criteria of an MPN*with eosinophilia associated with FIP1L1-PDGFRA |
| A myeloid or lymphoid neoplasm, usually with prominent eosinophilia |
| and |
| Presence of a FIP1L1-PDGFRA fusion gene or a variant fusion gene with rearrangement of PDGFRA† |
| Diagnostic criteria for myeloid/lymphoid neoplasms associated with ETV6-PDGFRB fusion gene or other rearrangement of PDGFRB‡ |
| A myeloid or lymphoid neoplasm, often with prominent eosinophilia and sometimes with neutrophilia or monocytosis |
| and |
| Presence of t(5;12)(q31∼q33;p12) or a variant translocation¶ or demonstration of an ETV6-PDGFRB fusion gene or rearrangement of PDGFRB |
| Diagnostic criteria of MPN or acute leukemia associated with FGFR1 rearrangement |
| A myeloproliferative or myelodysplastic/myeloproliferative neoplasm with prominent eosinophilia, and sometimes with neutrophilia or monocytosis |
| or |
| Acute myeloid leukemia or precursor T-cell or precursor B-cell lymphoblastic leukemia/lymphoma or mixed phenotype acute leukemia (usually associated with peripheral blood or BM eosinophilia) |
| and |
| Presence of t(8;13)(p11;q12) or a variant translocation leading to FGFR1 rearrangement demonstrated in myeloid cells, lymphoblasts, or both |
| Diagnostic criteria for myeloid/lymphoid neoplasms with PCM1-JAK2 |
| A myeloid or lymphoid neoplasm, often with prominent eosinophilia |
| and |
| Presence of t(8;9)(p22;p24.1) or a variant translocation leading to JAK2 rearrangement§ |
| CEL, NOS |
| There is eosinophilia (eosinophil count >1.5 × 109/L) |
| Not meeting WHO criteria for BCR-ABL1-positive chronic myeloid leukemia, PV, ET, PMF, CNL, CMML, or atypical CML |
| No rearrangement of PDGFRA, PDGFRB, or FGFR1; no PCM1-JAK2, ETV6-JAK2, or BCR-JAK2 fusion gene |
| The blast cell count in the peripheral blood and BM is less than 20%, and inv(16)(p13.1q22), t(16;16)(p13;q22) and other diagnostic features of AML are absent |
| There is a clonal cytogenetic or molecular genetic abnormality, or blast cells are ≥2% in the peripheral blood or >5% in the BM |
| Idiopathic HES |
| Exclusion of the following: |
| Reactive eosinophilia |
| Lymphocyte-variant HE (cytokine-producing, immunophenotypically-aberrant T-cell population) |
| CEL, NOS |
| WHO-defined myeloid malignancies associated eosinophilia (eg, MDS, MPNs, MDS/MPNs, or AML) |
| Eosinophilia-associated MPNs or AML/ALL with rearrangements of PDGFRA, PDGFRB, or FGFR1 or with PCM1-JAK2 |
| The absolute eosinophil count of >1.5×109/L must persist for at least 6 mo, and tissue damage must be present. If there is no tissue damage, idiopathic HES is the preferred diagnosis. |
| Myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, or FGFR1, or with PCM1-JAK2 . |
|---|
| Diagnostic criteria of an MPN*with eosinophilia associated with FIP1L1-PDGFRA |
| A myeloid or lymphoid neoplasm, usually with prominent eosinophilia |
| and |
| Presence of a FIP1L1-PDGFRA fusion gene or a variant fusion gene with rearrangement of PDGFRA† |
| Diagnostic criteria for myeloid/lymphoid neoplasms associated with ETV6-PDGFRB fusion gene or other rearrangement of PDGFRB‡ |
| A myeloid or lymphoid neoplasm, often with prominent eosinophilia and sometimes with neutrophilia or monocytosis |
| and |
| Presence of t(5;12)(q31∼q33;p12) or a variant translocation¶ or demonstration of an ETV6-PDGFRB fusion gene or rearrangement of PDGFRB |
| Diagnostic criteria of MPN or acute leukemia associated with FGFR1 rearrangement |
| A myeloproliferative or myelodysplastic/myeloproliferative neoplasm with prominent eosinophilia, and sometimes with neutrophilia or monocytosis |
| or |
| Acute myeloid leukemia or precursor T-cell or precursor B-cell lymphoblastic leukemia/lymphoma or mixed phenotype acute leukemia (usually associated with peripheral blood or BM eosinophilia) |
| and |
| Presence of t(8;13)(p11;q12) or a variant translocation leading to FGFR1 rearrangement demonstrated in myeloid cells, lymphoblasts, or both |
| Diagnostic criteria for myeloid/lymphoid neoplasms with PCM1-JAK2 |
| A myeloid or lymphoid neoplasm, often with prominent eosinophilia |
| and |
| Presence of t(8;9)(p22;p24.1) or a variant translocation leading to JAK2 rearrangement§ |
| CEL, NOS |
| There is eosinophilia (eosinophil count >1.5 × 109/L) |
| Not meeting WHO criteria for BCR-ABL1-positive chronic myeloid leukemia, PV, ET, PMF, CNL, CMML, or atypical CML |
| No rearrangement of PDGFRA, PDGFRB, or FGFR1; no PCM1-JAK2, ETV6-JAK2, or BCR-JAK2 fusion gene |
| The blast cell count in the peripheral blood and BM is less than 20%, and inv(16)(p13.1q22), t(16;16)(p13;q22) and other diagnostic features of AML are absent |
| There is a clonal cytogenetic or molecular genetic abnormality, or blast cells are ≥2% in the peripheral blood or >5% in the BM |
| Idiopathic HES |
| Exclusion of the following: |
| Reactive eosinophilia |
| Lymphocyte-variant HE (cytokine-producing, immunophenotypically-aberrant T-cell population) |
| CEL, NOS |
| WHO-defined myeloid malignancies associated eosinophilia (eg, MDS, MPNs, MDS/MPNs, or AML) |
| Eosinophilia-associated MPNs or AML/ALL with rearrangements of PDGFRA, PDGFRB, or FGFR1 or with PCM1-JAK2 |
| The absolute eosinophil count of >1.5×109/L must persist for at least 6 mo, and tissue damage must be present. If there is no tissue damage, idiopathic HES is the preferred diagnosis. |
Patients presenting with myeloproliferative neoplasm, AML, or lymphoblastic leukemia/lymphoma with eosinophilia and a FIP1L1-PDGFRA fusion gene are also assigned to this category.
If appropriate molecular analysis is not available, this diagnosis should be suspected if there is a Ph-chromosome–negative MPN with the hematologic features of chronic eosinophilic leukemia associated with splenomegaly, a marked elevation of serum vitamin B12, elevation of serum tryptase, and increased BM mast cells.
Cases with fusion genes typically associated only with BCR-ABL1-like B-lineage ALL are specifically excluded.
Because t(5;12)(q31∼q33;p12) does not always lead to an ETV6-PDGFRB fusion gene, molecular confirmation is highly desirable. If molecular analysis is not available, this diagnosis should be suspected if there is a Ph-chromosome–negative MPN associated with eosinophilia and with a translocation with a 5q31∼33 breakpoint.
Other variants giving rise to a fusion gene between JAK2 and an alternative partner include ETV6-JAK2 [t(9;12)(p24.1;p13.2)] or BCR-JAK2 [t(9;22)(p24.1;q11.2)].
Diagnosis
The differentiation of reactive vs clonal eosinophilia relies on a combined evaluation of blood counts/morphology, serum chemistry, organ damage (primarily physical examination, chest X-ray, ultrasound endoscopy, computed tomography/magnetic resonance imaging), BM morphology/immunohistochemistry, and molecular analyses including fluorescent-in situ-hybridization (FISH), polymerase chain reaction from RNA/cDNA and DNA with or without sequencing, and flow cytometry.5
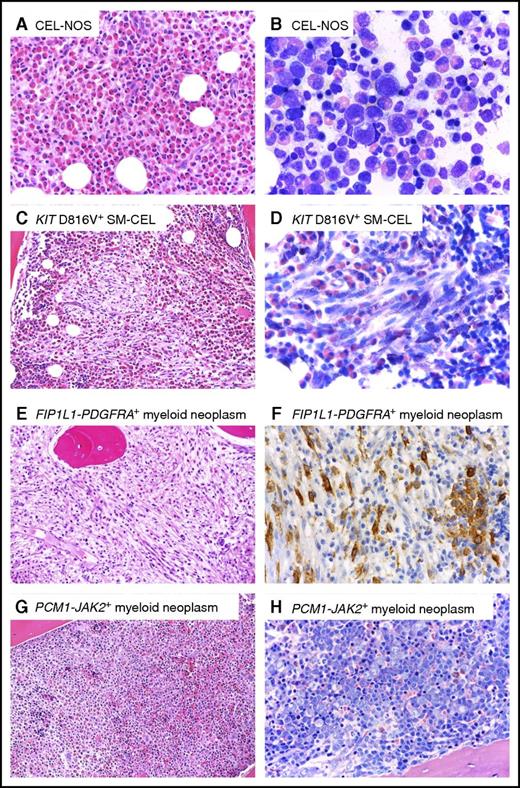
Peripheral blood smears should be screened for blasts, monocytosis, basophilia, left-shifted leukocytosis, dysplasia, or leukoerythroblastosis, which have been associated with the MPN or MDS/MPN phenotypes of rearranged PDGFRA/B, FGFR1, and JAK2.6 Importantly, eosinophilia is not an invariable feature of TK-rearranged myeloid/lymphoid neoplasms. Dysplastic or immature morphology of eosinophils or BM fibrosis may hint toward clonal eosinophilia, but cannot always be relied on to distinguish clonal vs reactive processes.5,6,12 Elevated serum tryptase and/or vitamin B12 levels may be observed with myeloid neoplasms, particularly in cases with PDGFRA or PDGFRB fusion genes.5,13 Increased serum tryptase levels can reflect the presence of loose clusters of atypical mast cells in the BM of these patients.14 However, the finding of a serum tryptase level higher than 100 ng/mL and elevated alkaline phosphatase ± splenomegaly and/or ascites is highly suggestive of systemic mastocytosis (SM), which is characterized by the KIT D816V mutation in >90% of cases and dense mast cell aggregates in the BM or other extracutaneous organs.15,16 In approximately 20% to 30% of cases, eosinophilia can accompany SM as an associated hematologic neoplasm. This has been variably referred to as SM-eo or SM with chronic eosinophilic leukemia (SM-CEL). The latter term may be preferable, as the KIT D816V mutation has been demonstrated in eosinophils, indicating they are part of a multilineage, neoplastic clone.17,18 Somatic missense mutations in the ethanolamine kinase gene (ETNK1) appear to be enriched in the subgroup of patients with SM with eosinophilia.19 Figure 2 demonstrates the unique histomorphologic BM features of 4 different eosinophilia-associated myeloid neoplasms, including SM-CEL.
Representative BM histomorphology of different myeloid neoplasms with eosinophilia. (A-B) CEL, NOS. (A) Hematoxylin and eosin (H&E). Histology of the BM shows an extremely hypercellular marrow with packed infiltrates of eosinophils. Normal blood cell precursors are absent. (B) Wright-Giemsa. Cytomorphological aspects on a BM smear also exhibit an abundance of eosinophils at all stages of maturation. Note that significant cellular atypia is absent. (C-D) KIT D816V-positive systemic mastocytosis with associated chronic eosinophilic leukemia (SM-CEL). (C) H&E. BM histology reveals a markedly hypercellular marrow with a massive increase in eosinophils. Centrally located in the marrow space, a compact infiltrate consisting of pale, mostly spindle-shaped cells is easily detected (left). (D) Higher magnification and Wright-Giemsa stain reveals that the spindle-shaped cells contain metachromatic granules, although in lesser number than normal mast cells. The presence of compact infiltrates mainly consisting of spindle-shaped cells allows prompt diagnosis of a systemic mastocytosis. (E-F) FIP1L1-PDGFRA-positive myeloid neoplasm. (E) H&E-stained BM shows massive hypercellularity with increase in eosinophils, which form narrow rows as an indirect sign of fibrosis. Normal blood cell precursors, in particular megakaryocytes, are missing. (F) ABC method. Anti-CD117 immunohistochemistry reveals an increase in spindle-shaped, mostly loosely scattered mast cells, with expression of CD117 (KIT). Note a small cohesive group of round mast cells on the right not sufficient for a diagnosis of systemic mastocytosis to be established. Mast cells display aberrant expression of CD25 (not depicted), which is seen outside the setting of mastocytosis only in FIP1L1-PDGFRA-positive myeloid neoplasms. (G-H) PCM1-JAK2-positive myeloid neoplasm. (G) H&E. (H) Wright-Giemsa. Extremely hypercellular BM with subtotal depletion of fat cells and massive increase in erythroblasts forming irregular giant erythra filling whole marrow spaces, and thus exceeding the size seen in any other myeloid neoplasm or reactive states. Images courtesy of Professor Hans-Peter Horny.
Representative BM histomorphology of different myeloid neoplasms with eosinophilia. (A-B) CEL, NOS. (A) Hematoxylin and eosin (H&E). Histology of the BM shows an extremely hypercellular marrow with packed infiltrates of eosinophils. Normal blood cell precursors are absent. (B) Wright-Giemsa. Cytomorphological aspects on a BM smear also exhibit an abundance of eosinophils at all stages of maturation. Note that significant cellular atypia is absent. (C-D) KIT D816V-positive systemic mastocytosis with associated chronic eosinophilic leukemia (SM-CEL). (C) H&E. BM histology reveals a markedly hypercellular marrow with a massive increase in eosinophils. Centrally located in the marrow space, a compact infiltrate consisting of pale, mostly spindle-shaped cells is easily detected (left). (D) Higher magnification and Wright-Giemsa stain reveals that the spindle-shaped cells contain metachromatic granules, although in lesser number than normal mast cells. The presence of compact infiltrates mainly consisting of spindle-shaped cells allows prompt diagnosis of a systemic mastocytosis. (E-F) FIP1L1-PDGFRA-positive myeloid neoplasm. (E) H&E-stained BM shows massive hypercellularity with increase in eosinophils, which form narrow rows as an indirect sign of fibrosis. Normal blood cell precursors, in particular megakaryocytes, are missing. (F) ABC method. Anti-CD117 immunohistochemistry reveals an increase in spindle-shaped, mostly loosely scattered mast cells, with expression of CD117 (KIT). Note a small cohesive group of round mast cells on the right not sufficient for a diagnosis of systemic mastocytosis to be established. Mast cells display aberrant expression of CD25 (not depicted), which is seen outside the setting of mastocytosis only in FIP1L1-PDGFRA-positive myeloid neoplasms. (G-H) PCM1-JAK2-positive myeloid neoplasm. (G) H&E. (H) Wright-Giemsa. Extremely hypercellular BM with subtotal depletion of fat cells and massive increase in erythroblasts forming irregular giant erythra filling whole marrow spaces, and thus exceeding the size seen in any other myeloid neoplasm or reactive states. Images courtesy of Professor Hans-Peter Horny.
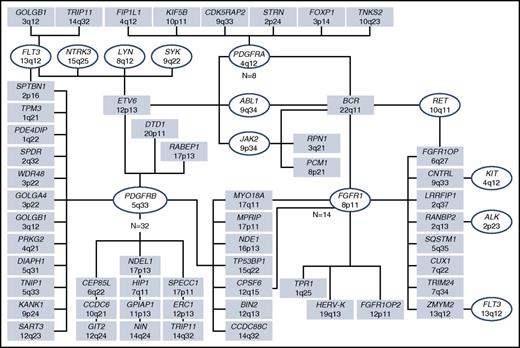
FISH for the CHIC2 deletion and/or reverse transcription polymerase chain reaction (RT-PCR) is used to detect the cytogenetically occult FIP1L1-PDGFRA fusion. Evidence for an alternative PDGFRA, PDGFRB, FGFR1, or JAK2 fusion gene can be inferred by cytogenetically visible breakpoints: rearrangement of 4q12 (PDGFRA), 5q31∼33 (PDGFRB), 8p11∼12 (FGFR1), and 9p24 (JAK2). Rearrangements of ABL1 and FLT3, which are also associated with eosinophilia, can similarly be inferred by their respective breakpoints, 9q34 and 13q12. More than 70 fusion genes associated with myeloid/lymphoid neoplasms with eosinophilia have been reported to date (Figure 3).
TK fusion genes in myeloid/lymphoid neoplasms with eosinophilia. As of October 2016, 72 tyrosine kinase fusion genes have been described. In conjunction with cytogenetic/fluorescence in situ hybridization and genetic testing, morphologic evaluation and flow cytometric immunophenotyping is required to identify whether such molecular abnormalities are associated with an acute or chronic myeloid neoplasm and/or a B- or T-cell lymphoblastic leukemia/lymphoma. Figure courtesy of Professor Nicholas C. P. Cross.
TK fusion genes in myeloid/lymphoid neoplasms with eosinophilia. As of October 2016, 72 tyrosine kinase fusion genes have been described. In conjunction with cytogenetic/fluorescence in situ hybridization and genetic testing, morphologic evaluation and flow cytometric immunophenotyping is required to identify whether such molecular abnormalities are associated with an acute or chronic myeloid neoplasm and/or a B- or T-cell lymphoblastic leukemia/lymphoma. Figure courtesy of Professor Nicholas C. P. Cross.
Although the classification of myeloid neoplasms with eosinophilia has increasingly relied on molecular markers, diagnosis must still be anchored to a combination of histomorphology and clinical and laboratory criteria. In fact, none of the molecular markers is linked to a singular phenotype. For example, FIP1L1-PDGFRA may rarely be associated with T-lymphoblastic lymphoma/leukemia, despite usually presenting as a chronic myeloid neoplasm with eosinophilia; similarly, PDGFRB rearrangements can lead to a spectrum of myeloid neoplasms. Fusion genes involving FGFR1, JAK2, FLT3, and ABL1 are each associated with diverse presentations, including B- or T-cell lymphoid leukemias/lymphomas or chronic or de novo/secondary acute myeloid leukemias with or without eosinophilia. It is likely that host factors (such as patients’ germline susceptibility alleles) and disease factors (eg, the tumor landscape of acquired somatic mutations and subclones) contribute to disease heterogeneity. Although the WHO classification provides a useful platform for stratifying these eosinophilic neoplasms, it is incumbent on the treating physician to take into account both clinicopathologic and molecular criteria to refine disease diagnosis and prognosis and to establish the appropriate treatment plan.
FIP1L1-PDGFRA and other PDGFRA fusion genes
In 2001 to 2002, several reports described rapid and complete hematologic remissions with empiric use of imatinib in patients diagnosed with HES or CEL.20-22 The therapeutic target of imatinib was identified as the chimeric oncoprotein encoded by FIP1L1-PDGFRA.23,24 The fusion disrupts the autoinhibitory juxtamembrane domain of PDGFRA, resulting in constitutive activation of the TK. FIP1L1-PDGFRA is generated from a submicroscopic 800-kb interstitial deletion on chromosome 4, del(4)(q12q12).23,24 The deleted segment contains the CHIC2 gene, which is the basis for the commonly used FISH test used to diagnose FIP1L1-PDGFRA-positive disease.25 However, because of the rare possibility of complex rearrangements or variant FIP1L1 breakpoints, FISH and RT-PCR should be used together in patients with a high index of suspicion for FIP1L1-PDGFRA, but in whom 1 of these diagnostic tests returns a negative result.26 Although FIP1L1-PDGFRA-positive disease usually presents as a chronic MPN with eosinophilia, it may be diagnosed in the blastic phase of a MPN or as an eosinophilia-associated AML or T-cell lymphoblastic lymphoma.27 The basis for the striking male predominance among FIP1L1-PDGFRA-positive patients has not been elucidated.
Seven additional fusion partners of PDGFRA have been described, including BCR, ETV6, KIF5B, CDK5RAP2, STRN, TNKS2, and FOXP1. In addition, point mutations in PDGFRA have been identified in patients with a diagnosis of HES.28 Although differences exist in their transforming ability, some mutants induced a leukemia-like disease in mice that was responsive to imatinib.
Several studies have corroborated the high rates of complete hematologic remission (CHR) and complete molecular remission (CMR) with imatinib therapy of 100 to 400 mg daily in FIP1L1-PDGFRA-positive MPN-eo (Table 3).29-35 Although the optimal induction dose is not defined, 100 mg daily is sufficient to elicit CMRs in a majority of patients. Maintenance doses as low as 100 to 200 mg weekly can sustain CMRs.36 The durability of these excellent outcomes has prompted investigators to assess the feasibility of stopping imatinib in those patients. Outcomes data are based on limited numbers of patients and reveal substantial variability in relapse-free survival.26,27,32,34,35,37,38 It is currently unknown what factors predict long-term CMR in these individuals, and therefore, discontinuation of imatinib should generally be undertaken in the context of clinical trials or registries.
Series evaluating imatinib in FIP1L1-PDGFRA-positive myeloid/lymphoid neoplasms
| Reference . | n . | CHR (%) . | CMR (%) . | Follow-up in months, median (range) . | Resistance . | Deaths . |
|---|---|---|---|---|---|---|
| Baccarani et al Haematologica, 200730 | 27 | 100 | 100 | 25 (15-60) | — | — |
| Klion et al J Allergy Clin Immunol., 200931 | 17 | N/A | 88 | N/A | — | — |
| Helbig et al Hematol Oncol., 201033 | 16 | 100 | 100 | 36 (2-59) | — | — |
| Pardanani et al Leukemia, 201234 | 18 | 94 | 100 | 73 | 1 | 2 |
| Legrand et al Medicine, 201335 | 44 | 100 | 95 | 52 (1-100) | — | 1 |
| German Registry on Disorders of Eosinophils and Mast Cells (unpublished) | 64 | 100 | 90 | 77 (2-129) | 2 | 4 |
| Reference . | n . | CHR (%) . | CMR (%) . | Follow-up in months, median (range) . | Resistance . | Deaths . |
|---|---|---|---|---|---|---|
| Baccarani et al Haematologica, 200730 | 27 | 100 | 100 | 25 (15-60) | — | — |
| Klion et al J Allergy Clin Immunol., 200931 | 17 | N/A | 88 | N/A | — | — |
| Helbig et al Hematol Oncol., 201033 | 16 | 100 | 100 | 36 (2-59) | — | — |
| Pardanani et al Leukemia, 201234 | 18 | 94 | 100 | 73 | 1 | 2 |
| Legrand et al Medicine, 201335 | 44 | 100 | 95 | 52 (1-100) | — | 1 |
| German Registry on Disorders of Eosinophils and Mast Cells (unpublished) | 64 | 100 | 90 | 77 (2-129) | 2 | 4 |
Resistance to imatinib is rare in FIP1L1-PDGFRA-positive disease. Almost all cases relate to acquisition of the T674I mutation within the ATP-binding domain of PDGFRα,23,39-41 which is analogous to the BCR-ABL1 T315I mutation in chronic myeloid leukemia (CML) and confers pan-resistance to multiple TK inhibitors (TKI), except ponatinib.42 One patient with the FIP1L1-PDGFRA T674I mutation in blast phase responded briefly to sorafenib, but was followed by rapid emergence of a pan-resistant FIP1L1-PDGFRA D842V mutant and death of the patient.41 Despite in vitro nanomolar activity of sorafenib, midostaurin, or nilotinib against the T674I mutant,43-46 these drugs have shown limited clinical activity.47 Although drugs with more potent activity against T674I or D842V are in clinical development, only allogeneic hematopoietic stem cell transplantation (HSCT) may be able to extend disease-free survival.
PDGFRB fusion genes
Since the initial identification of ETV6-PDGFRB in 1994,48 more than 30 additional fusion partners of PDGFRB have been characterized. Most patients with these fusions have been described as having a chronic MPN, MDS/MPN overlap neoplasm (eg, chronic myelomonocytic leukemia [CMML], atypical chronic myeloid leukemia [CML], juvenile myelomonocytic leukemia [JMML]), or rarely myeloid blast phase/secondary AML. Eosinophilia is a common, but not invariable feature of cases with PDGFRB fusion genes. FISH or RT-PCR is necessary to prove involvement of the PDGFRB gene, as several genes encoding eosinophilopoietic cytokines (eg, IL-3, IL-5, GM-CSF) reside in the 5q31∼33 region.
Several common themes are relevant to the biology of PDGFRB fusion genes.49,50 They are in-frame and share the same molecular breakpoint, and the PDGFRβ TK domain is preserved and fused to an N-terminal protein in all the chimeric oncoproteins. In addition, the amino terminal partner protein always contains dimerization/oligomerization motifs that can mimic receptor dimerization and activation in the absence of PDGFRB ligand. The in vitro and in vivo transforming activity depends on the presence of a catalytic domain of PDGFRβ and the dimerization motifs of the partner protein.
The standard imatinib dose used for patients with chronic phase PDGFRB-rearranged eosinophilic MPNs is 100 to 400 mg daily.51 Cheah et al reported the long-term follow-up (median, 10.2 years) of 26 PDGFRB-rearranged patients treated with imatinib for a median duration of 6.6 years.52 Imatinib demonstrated a 96% response rate, a 6-year progression-free survival rate of 88%, and a 10-year overall survival rate of 90%. None of the patients who achieved a complete cytogenetic or molecular remission lost their response or exhibited progression to blast phase.
FGFR1 fusion genes
FGFR1 fusion genes are associated with a disease entity initially referred to as the 8p11 myeloproliferative syndrome or stem cell leukemia/lymphoma.9,53 Fourteen different fusion genes have now been described. The 3 most common reciprocal translocations include t(8;13)(p11;q12), t(8;9)(p11;q33), and t(6;8)(q27;p11), resulting in fusions of ZMYM2, CNTRL, and FGFR1OP, respectively, to FGFR1.54-56 The transforming activity of the various FGFR1 fusion proteins has been demonstrated by conversion of the IL-3-dependent Ba/F3 cell line to growth factor independence and induction of a myeloproliferative neoplasm in murine models.57,58
The clinical/laboratory characteristics typically reflect features of chronic myeloid neoplasms and variable eosinophilia.9 Patients may also present as de novo AML without an antecedent MPN. There is a high incidence of T-lymphoblastic lymphomas, particularly in association with a t(8;13) and a ZMYM2-FGFR1 fusion gene,9,53,54 which may occur at diagnosis or during the course of disease, reflecting a myeloid/lymphoid stem cell origin. The clinical course is aggressive as a result of rapid progression to blast phase/secondary acute leukemia, usually of myeloid phenotype, less commonly B-ALL, within 1 or 2 years of diagnosis. The variability in the clinical presentation may be a result of specific moieties of the partner genes and signaling via different intracellular pathways.59 The t(8;22) is often associated with a clinical and hematologic picture very similar to that seen in BCR-ABL1-positive CML with basophilia,58,60 whereas thrombocytopenia and monocytosis resembling CMML are more frequently present in t(6;8)56 and t(8;9).55,61 The t(6;8) may also present with a PV-like disease,62 and eosinophilia may be absent in t(6;8) and t(8;22).
In the absence of allogeneic HSCT, which is the only potentially curative treatment option for eligible patients, chemotherapy resistance and rapid disease progression are the usual outcome. FGFR1 fusions are generally resistant to imatinib, nilotinib, and dasatinib; however, midostaurin elicited a hematologic response in 1 patient.63
Ponatinib can inhibit proliferation and/or induce apoptosis of FGFR1-transformed cells and reduce colony growth of cells from patients with FGFR1-rearranged disease.64 Ponatinib was used as monotherapy and subsequently combined with intensive ALL-type induction chemotherapy (eg, hyperCVAD [alternating courses of chemotherapy with course A (cyclophosphamide, vincristine, doxorubicin, and dexamethasone) and course B (cytarabine and methotrexate)]) as a bridge to allogeneic HSCT in a patient with a BCR-FGFR1-positive trilineage mixed-phenotype acute leukemia.65 A.R. and colleagues evaluated the efficacy of standard-dose ponatinib in 7 FGFR1-rearranged patients.66 There was either progressive disease or no evidence for sustained hematologic or cytogenetic response. There was similarly no evidence for sustained complete remissions with intensive chemotherapy. Four patients ultimately underwent allogeneic HSCT and were reported in CMR and alive after a median time of 19 months (range, 8-36 months) after diagnosis and 13 months (range, 4-29 months) after allogeneic HSCT. These data support the notion that allogeneic HSCT is currently the only option to achieve long-term remission, or possibly cure, in patients with FGFR1 fusion genes.
JAK2 fusion genes
The most frequent JAK2 fusion gene is PCM1-JAK2 as a consequence of a t(8;9)(p22;p24),67-69 whereas ETV6-JAK2 [t(9;12)(p24;p13)] and BCR-JAK2 [t(9;22)(p24;q11)] fusion genes have been reported in only a few patients.70-74 To date, murine models have only been established for ETV6-JAK2, which showed the induction of a fatal mixed myelo- and/or T-cell lymphoproliferative disorder and potential susceptibility to JAK inhibitors.75,76
The phenotype and clinical course of JAK2-rearranged patients resemble leukemias associated with other TK fusion genes (eg, BCR-ABL1-positive CML/ALL), rather than classical Philadelphia-chromosome-negative MPNs with the JAK2 V617F mutation.71 PCM1-JAK2-positive patients usually present with features of a chronic myeloid neoplasm (eg, MPN or MDS/MPN) and are frequently (50% to 70%) associated with eosinophilia and/or BM fibrosis.67,71 The clinical course of JAK2-rearranged neoplasms is aggressive, with rapid progression from chronic phase disease to AML, and more rarely to lymphoid blast phase, which has been associated with acquisition of biallelic IKZF1 alteration as well as EBF1 and CDKN2A/B codeletions.77 Patients may also present as de novo acute leukemia, with myeloid being more common than lymphoid origin.73 The involvement of both myeloid and lymphoid lineages demonstrates that PCM1-JAK2-positive neoplasms originate from an early pluripotent hematopoietic progenitor or stem cell. Because of the clear analogies with the multilineage neoplasms associated with rearrangements of PDGFRA/B or FGFR1, it was therefore suggested that PCM1-JAK2-positive neoplasms be recognized as a provisional entity in the new WHO 2016 classification. ETV6-JAK2 and BCR-JAK2 may be considered variants of PCM1-JAK2; the small number of ETV6-JAK2 and BCR-JAK2 cases have presented as myeloid neoplasms with variable presence of eosinophilia or features of BCR-ABL1-like ALL.
Allogeneic HSCT was performed in 5 (38%) of 13 patients with PCM1-JAK2 fusions who were younger than 55 years either in first chronic phase or in first remission after intensive chemotherapy of acute leukemia.71 At the time of reporting, 4 patients were alive and progression-free at a median time of 33 months after transplant. One patient died after 5 months because of graft failure. Twelve patients did not receive an allogeneic HSCT, of whom 5 were alive and progression-free. Seven nontransplanted patients died at a median time of 12 months, with only 1 death not being disease-related. Overall, the data suggest that the PCM1-JAK2 fusion is associated with a poor prognosis, and early allogeneic HSCT should be considered for eligible patients.
After the success of imatinib in patients with ABL1 and PDGFR fusion genes, it was hypothesized that JAK inhibitors such as ruxolitinib may be capable of inducing similar remissions in patients with JAK2 fusion genes. A patient with CEL who was treated with ruxolitinib 15 months after diagnosis achieved a complete cytogenetic response and was alive at 30 months.78 Two patients with CEL exhibited complete hematologic and cytogenetic remissions, as well as marked reduction of PCM1-JAK2 transcripts, at 33 and 46 months after the start of ruxolitinib therapy at the time of publication.79,80 Schwaab et al treated 2 male patients with chronic myeloid neoplasms and a PCM1-JAK2 or a BCR-JAK2 fusion gene, respectively, with ruxolitinib.81 After 12 months of treatment, both patients achieved a complete clinical, hematologic, and cytogenetic response. However, remission in both patients was only short-term, with relapse occurring after 18 and 24 months, respectively, making allogeneic HSCT indispensable in both cases.81 Larger series are needed to assess the value of the long-term efficacy of ruxolitinib in JAK2-rearranged cases. Their optimal use may be for disease cytoreduction before allogeneic HSCT.
ETV6-FLT3 and other FLT3 fusion genes
Although internal tandem duplications and point mutations in FLT3 are well described in AML,82 fusion genes involving FLT3 are extremely rare. The majority of cases with a FLT3 rearrangement have involved ETV6-FLT3, with 6 cases reported since 2006.83-88 The ETV6-FLT3 fusion causes IL-3-independent transformation of Ba/F3 murine hematopoietic cells.83 It is hypothesized that the helix–loop–helix oligomerization domain of ETV6-FLT3 promotes ligand-independent dimerization, resulting in constitutive TK activity.
The phenotype in all FLT3-rearranged cases was a myeloid and/or lymphoid neoplasm with eosinophilia, consistent with the WHO category of CEL, NOS. Four of the 6 ETV6-FLT3 cases exhibited an associated diagnosis of peripheral T-cell lymphoma or T-lymphoblastic lymphoma.85-87 In addition to ETV6-FLT3, there has been 1 case report describing a SPTBN1-FLT3 translocation in a patient presenting with a phenotype of CML-like disease with eosinophilia,88 and another report describing a patient with an eosinophilia-associated MPN and T-cell lymphoma resulting from a GOLGB1-FLT3 fusion.89 In addition, we have recently identified a TRIP11-FLT3 fusion gene in a patient with CEL, NOS that rapidly transformed to a fatal T-cell acute lymphoblastic leukemia/lymphoma (J.G., unpublished observation).
Similar to the experience with AML, multikinase inhibitors with anti-FLT3 activity have demonstrated relatively modest activity and limited durability in patients with FLT3 rearrangements. In 2 cases of ETV6-FLT3-positive CEL, sunitinib elicited complete hematologic and/or cytogenetic responses, but were short-lived as a result of rapid emergence of disease progression.85 More promising, Falchi et al described a patient with ETV6-FLT3-positive CEL who achieved a complete hematologic and partial cytogenetic remission after only 1.5 months of treatment with sorafenib.84 The patient subsequently underwent matched sibling donor allogeneic HSCT and was alive and in remission 11 months after initial diagnosis. These studies suggest a potential role of FLT3 inhibitors, but monotherapy is unlikely to elicit long-term disease control and should instead be used as a bridge to allogeneic HSCT.
ETV6-ABL1 fusion gene
The ETV6-ABL1 fusion gene results from a t(9;12)(q34;p13) or complex rearrangements.90-93 Importantly, the creation of the ETV6-ABL1 fusion requires at least 3 chromosomal breaks, usually a cryptic insertion, to generate an in-frame fusion gene.93,94 Routine karyotyping is therefore usually inconclusive, and FISH can miss the small insertions. Only a combination of ETV6 and ABL1 probes, targeted RT-PCR, or RNAseq can reliably identify the fusion.94
In children, the clinical phenotype of ETV6-ABL1-positive patients is predominantly de novo ALL; however, similar to all other TK fusion genes in adults, various AML and chronic myeloid/lymphoid phenotypes have been described. Zaliova et al recently reported on the incidence, clinical features, and genetics of ETV6-ABL1 leukemias in 44 cases.94 Overall, these cases are strongly reminiscent of BCR-ABL1-positive neoplasms. Phenotypes included ALL (n = 22: children, n = 13; adults, n = 9) and myeloid neoplasms (n = 22: MPN, n = 18; AML, n = 4). Of interest, eosinophilia was present in all MPN and AML cases, but only four of 13 ALL cases.94 In adults, the prognosis of acute leukemias is very poor, whereas patients with MPN have significantly better survival. Among those evaluable, more than 80% of patients died because of disease progression or relapse. In patients with MPN, 2 patients were treated with imatinib, 3 were treated sequentially with imatinib and nilotinib because of progressive disease, and all patients were alive at reporting. TKI were less effective in 5 patients in blast phase, with 4 patients having died, 3 within 1 year.
Blast phase disease
Careful attention should be paid to clinical situations in which eosinophilia is concurrently diagnosed with de novo or blast phase/secondary acute leukemias of myeloid or lymphoid phenotype, high-grade lymphomas (usually of T-cell subtype), or myeloid sarcomas because FIP1L1-PDGFRA and other TK fusion genes have been reported in numerous cases.27,95-97 We are aware of several cases in which a fusion gene has only come to attention after patients achieved clearance of blasts or remission of lymphomas with intensive chemotherapy but significant eosinophilia persisted. If performed, molecular analyses have shown that the genetic lesion is usually detectable in granulocytic cells, but also in blasts of myeloid or lymphoid origin, indicating a stem cell disorder with multilineage involvement and a disparate morphologic appearance in BM and lymph nodes.
In a case series of 17 patients with PDGFRA/B fusion gene-positive blast phase or sarcoma, 15 patients treated with imatinib monotherapy achieved durable complete hematologic and molecular remissions. Only 2 (12%) of 17 patients died after a median observation time of 65 months (range, 7-106).97 Two patients were initially treated with intensive chemotherapy instead of imatinib, but failed to achieve a complete remission; subsequent allogeneic HSCT was shortly followed by relapse within 3 months.97 The combination of imatinib and intensive chemotherapy, similar to that which has been undertaken with ALL induction regimens such as hyperCVAD in BCR-ABL1-positive ALL, merits further investigation. More data are also needed regarding the role of allogeneic HSCT, given the few numbers of PDGFRA/B-positive blast phase patients treated to date. Patients with ETV6-ABL1-positive de novo or secondary acute leukemias/blast phase have a very poor prognosis, despite TKI therapy, with median survival of approximately 1 year.94
CEL, NOS
An increase in blood or BM myeloblasts, and/or the presence of nonspecific cytogenetic abnormalities such as trisomy 8 or isochromosome 17 [i(17q)], comprise basic diagnostic criteria for CEL, NOS.11 With exception of PCM1-JAK2, the category of CEL, NOS, by default, also includes reciprocal translocations with rearrangement of JAK2, ABL1, and FLT3. Because of similar and frequently indistinguishable clinical characteristics and the involvement of a TK fusion gene, the inclusion of these abnormalities in the group of “myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB, FGFR1 or a PCM1-JAK2 fusion gene” should be considered.71 In a large series, of 496 unselected patients with eosinophils greater than 1.5 × 109/L (personal communication, T. Haferlach, Munich Leukemia Laboratory), blasts greater than 2% in PB and greater than 5% in BM were identified in 0 and 5 (1%) patients, respectively. Cytogenetic analysis was performed in 329 patients. Sixteen patients (4.9%) exhibited rearrangement of PDGFRA/B, and only 10 (3%) patients had cytogenetic aberrations such as other reciprocal translocations, trisomies, or a complex karyotype.
No standard of care exists for the treatment of CEL, NOS. Hydroxyurea may be useful to control leukocytosis, eosinophilia, and splenomegaly in selected patients. Interferon-α can produce hematologic and cytogenetic remissions in CEL patients refractory to other therapies including prednisone and/or hydroxyurea.98-100
A case series of 10 subjects with CEL, NOS illustrates the generally poor outcomes of these patients.101 The median overall survival of the cohort was 22 months, with 5 (50%) patients developing AML after a median of 20 months from diagnosis. Three of the remaining 5 patients died of active disease, and 2 patients exhibited longer-term remissions with allogeneic HSCT (n = 1) and the use of imatinib/hydroxyurea (n = 1). Similar to patients with some specific rearranged TK fusion genes (eg, FGFR1, JAK2), allogenic HSCT may provide the best chance of long-term survival.
Next-generation sequencing identifies clonality in HEUS /HES patients
Recently, several reports have identified 1 or more recurrent point mutations in patients undergoing diagnostic work-up of HEUS or HES. Schwaab et al screened 426 unselected samples from patients with eosinophilia for KIT D816V and JAK2 V617F who were initially referred for the screening of FIP1L1-PDGFRA.16 Overall, 14 (3%) patients tested positive for KIT D816V, and 17 patients (4%) for JAK2 V617F. Median survival was 26 and 41 months, respectively, and was significantly shorter for all patients if absolute eosinophilia was greater than 2 × 109/L (median survival, 20 months vs not reached; P = .002).16
Pardanani et al investigated 98 patients with HEUS or HES for molecular abnormalities.102 Eleven (11%) harbored a known or predicted pathogenetic mutation; each patient had a mutation in 1 of 7 genes (TET2, 3; ASXL1, 2; KIT, 2; IDH2, 1; JAK2, 1; SF3B1, 1; TP53, 1). In addition, 15% of patients harbored a variant of unknown significance in 1 of 6 genes (TET2, ASXL1, SETBP1, CALR, CEBPA, and CSF3R). No differences were observed regarding clinical characteristics or pattern of end-organ involvement. On multivariable analysis, age greater than 60 years, hemoglobin levels lower than 10 g/dL, cardiac involvement, and hepatosplenomegaly were associated with inferior OS and used for a prognostic model that differentiated a high- vs a low-risk group with 5-year survival rates of 62% vs 98% (P < .0001). Patients harboring mutations in both low- and high-risk subgroups exhibited inferior OS, although differences in OS within each risk category were not statistically significant. The authors concluded that the identified mutations may not represent true driver mutations for HEUS/HES, and that a reclassification as CEL, NOS may be premature.
Wang et al reported the identification of 1 or more mutations by next-generation sequencing (NGS) in 14 (28%) of 51 patients who carried a diagnosis of HES.103 Single gene mutations were found in 7 patients, and 2 or more gene mutations were found in the additional 7 patients. Mutations included ASXL1 (43%), TET2 (36%), EZH2 (29%), CBL (14%), SETBP1 (22%), and NOTCH1 (14%). The clinical characteristics and poor overall survival of this group of HES patients with positive NGS results were similar to a control group of 17 patients with CEL, NOS, whose median overall survival was 14.4 months. In contrast, the 37 patients with HES and negative NGS testing exhibited more favorable survival compared with the patients with CEL, NOS, or HES with positive NGS results.
The identification of disease-specific mutations primarily leads to a diagnosis of SM (KIT D816V, ETNK1) or PV/ET/MF (JAK2 V617F) associated with eosinophilia and a rationale for use of TKIs such as midostaurin (KIT D816V) or ruxolitinib (JAK2 V617F). Some of the recently identified point mutations may also lead to a diagnosis of CEL, NOS, but further functional analysis of the transforming capacity of the detected variants is needed.
Future directions
Several opportunities for progress exist in myeloid neoplasms with eosinophilia. For the initial diagnostic evaluation of HE, reliable antibodies for fluorescence-activated cell sorting or immunohistochemistry to establish the clonality of eosinophils are needed. For patients in whom a clonal eosinophilia is suspected, but conventional cytogenetics, FISH, or RT-PCR testing is unremarkable, we advocate NGS vis à vis myeloid mutation panels to establish not only whether pathogenetic variants are present but also whether there are actionable targets. RNA sequencing for the identification of cytogenetically silent fusion genes may be applicable in selected cases.
Large worldwide registries or prospective multicenter trials should be initiated, similar to the STIM studies in CML, to identify clinical and molecular markers that may facilitate the prediction of which patients with a PDGFRA/B fusion gene in CMR can stop imatinib. In contrast to outstanding responses with imatinib in patients with rearranged PDGFRA/B, other TKIs often exhibit less activity against other fusion genes (Table 4), highlighting the need for more potent and selective agents against JAK2 and FGFR1. Current and future treatment directions for CEL, NOS (and HES) include evaluation of JAK inhibitors, siglec 8 agonists, and IL-5 receptor antibody (benralizumab). The anti-IL5 antibody mepolizumab was recently approved for patients with severe eosinophilic asthma, but is currently only available on an investigational basis for patients with HES. Such anti-IL-5 approaches may also be useful for CEL, NOS. The adoption of multicenter study protocols with consensus response criteria and informative biologic correlates will help catalyze drug development for these rare diseases.
Currently known TK fusion genes associated with myeloid/lymphoid neoplasms with eosinophilia and potential activity of TK inhibitors
| TKgene . | Rearranged chromosomal band . | Known partner genes (at least) . | Most frequent partner gene . | Preferably used inhibitors . | Risk for resistance/ progression/death . | Efficacy . |
|---|---|---|---|---|---|---|
| PDGFRA | 4q12 | 8 | FIP1L1 | Imatinib | Very low | +++ |
| PDGFRB | 5q31∼33 | >30 | ETV6 | Imatinib | Very low | +++ |
| FGFR1 | 8p11∼12 | 14 | ZMYM2 | Ponatinib | High | + |
| JAK2 | 9p24 | 4 | PCM1 | Ruxolitinib | Variable | ++ |
| ABL1 | 9q33 | 2 | ETV6 | Imatinib | Variable | ++ |
| Nilotinib | ||||||
| Dasatinib | ||||||
| FLT3 | 13q12 | 3 | ETV6 | Sorafenib | High | + |
| Sunitinib | ||||||
| Midostaurin |
| TKgene . | Rearranged chromosomal band . | Known partner genes (at least) . | Most frequent partner gene . | Preferably used inhibitors . | Risk for resistance/ progression/death . | Efficacy . |
|---|---|---|---|---|---|---|
| PDGFRA | 4q12 | 8 | FIP1L1 | Imatinib | Very low | +++ |
| PDGFRB | 5q31∼33 | >30 | ETV6 | Imatinib | Very low | +++ |
| FGFR1 | 8p11∼12 | 14 | ZMYM2 | Ponatinib | High | + |
| JAK2 | 9p24 | 4 | PCM1 | Ruxolitinib | Variable | ++ |
| ABL1 | 9q33 | 2 | ETV6 | Imatinib | Variable | ++ |
| Nilotinib | ||||||
| Dasatinib | ||||||
| FLT3 | 13q12 | 3 | ETV6 | Sorafenib | High | + |
| Sunitinib | ||||||
| Midostaurin |
Acknowledgments
J.G. expresses gratitude to the Charles and Ann Johnson Foundation for their support. This work was supported by grants from Deutsche José Carreras Leukämie-Stiftung (R 13/05 to A.R.). We also wish to thank Professors Hans-Peter Horny and Nicholas C. P. Cross for their collaborations and provision of figures for this review.
Authorship
Contribution: A.R. and J.G. both contributed to the writing of the manuscript.
Conflict-of-interest disclosure: A.R. and J.G. have received honoraria, travel support, and research funding and have served on an advisory board for Novartis Pharmaceuticals, Inc.
Correspondence: Jason Gotlib, Stanford University School of Medicine/Stanford Cancer Institute, 875 Blake Wilbur Dr, Room 2324, Stanford, CA 94305-5821; e-mail: jason.gotlib@stanford.edu.