Key Points
Gene polymorphism of the immune response as CTLA4 was shown to impact CBT outcomes according to CBU genotype.
CTLA4-CBU genotype might be considered for CBU selection when >1 CBU meeting the current suggested selection criteria is available.
Abstract
We evaluated the impact of recipient and cord blood unit (CBU) genetic polymorphisms related to immune response on outcomes after unrelated cord blood transplantations (CBTs). Pretransplant DNA samples from 696 CBUs with malignant diseases were genotyped for NLRP1, NLRP2, NLRP3, TIRAP/Mal, IL10, REL, TNFRSF1B, and CTLA4. HLA compatibility was 6 of 6 in 10%, 5 of 6 in 39%, and ≥4 of 6 in 51% of transplants. Myeloablative conditioning was used in 80%, and in vivo T-cell depletion in 81%, of cases. The median number of total nucleated cells infused was 3.4 × 107/kg. In multivariable analysis, patients receiving CBUs with GG-CTLA4 genotype had poorer neutrophil recovery (hazard ratio [HR], 1.33; P = .02), increased nonrelapse mortality (NRM) (HR, 1.50; P < .01), and inferior disease-free survival (HR, 1.41; P = .02). We performed the same analysis in a more homogeneous subset of cohort 1 (cohort 2, n = 305) of patients who received transplants for acute leukemia, all given a myeloablative conditioning regimen, and with available allele HLA typing (HLA-A, -B, -C, and -DRB1). In this more homogeneous but smaller cohort, we were able to demonstrate that GG-CTLA4-CBU was associated with increased NRM (HR, 1.85; P = .01). Use of GG-CTLA4-CBU was associated with higher mortality after CBT, which may be a useful criterion for CBU selection, when multiple CBUs are available.
Introduction
Cord blood grafts from an unrelated donor have been frequently used in the absence of a HLA-matched related or unrelated hematopoietic stem cell (HSC) transplant (HSCT) donor. To date, >35 000 unrelated cord blood transplantations (CBTs) have been performed worldwide and >630 000 cord blood units (CBUs) are available for transplantation.1
CBT outcomes depend on characteristics of CBUs (mainly cell dose and number of HLA disparities) and other factors related to recipients, underlying disease, and transplantation technique (such as conditioning regimen and graft-versus-host disease [GVHD] prophylaxis).2-4 Identification of factors to improve outcomes is of paramount importance, and may impact the current criteria for CBU selection.
Recently, better HLA matching based on allele typing has been shown to improve outcomes after CBT.5 Also, it has been described that non-HLA factors of CBUs, such as matching of killer immunoglobulin-like receptor ligand or noninherited maternal antigens, may also affect outcomes after CBT and could be used as criteria for CBU selection.6,7
With the development of human genomics, several studies have attempted to correlate genetic polymorphisms of the innate and adaptive immune response to hematopoietic cell transplantation (HCT) outcomes.8-16 However, in the CBT setting, only 1 study including a small and heterogeneous group of 115 CBT recipients has been described. In that study, pairs of genotyped samples (recipients and donors) for tumor necrosis factor α and interleukin 10, as well as for minor histocompatibility antigen, HY, histocompatibility antigen 1, and CD31 codon 125 were evaluated, and no significant association between polymorphisms and transplant outcomes was observed.17
Therefore, we conducted a retrospective cohort registry, based with the aim of studying the influence of genetic polymorphisms of CBU on transplant outcomes, namely myeloid engraftment, acute and chronic GVHD, nonrelapse mortality (NRM), relapse, and survival for patients with hematological malignancies. We selected 8 candidate genes (NLRP1, NLRP2, NLRP3, TIRAP/Mal, IL10, REL, TNFRSF1B, and CTLA4), which are genes encoding main cytokines and other immune proteins involved in the development of innate and adaptive immunological responses that have been associated with outcomes after HCT in previous studies. Tirap/mal mutation has been associated with NRM, whereas IL10 and TNFRSF1B have been associated with GVHD and survival. The NALP genes (NLRP1, NLRP2, NLRP3) have been associated with fungal infection and, as CTLA4, have also been associated with relapse and mortality. The association of CTLA4 with GVHD has also been previously observed.18-35 REL, a subunit of NF-κB, has been implicated with the inflammatory response in autoimmune disease and septic shock outcomes but, to our knowledge, our study is the first to analyze REL in the transplant setting.36,37
Patients and methods
Study design
This is a retrospective cohort study performed in collaboration with Eurocord, Cellular Therapy and Immunobiology Working Party of the European Society for Blood and Marrow Transplantation (EBMT-CTIWP), NetCord and Ribeirão Preto School of Medicine of São Paulo University (FMRP-USP).
Inclusion criteria were: (1) availability of CBU samples used in the CBT; (2) recipients of unrelated single CBT with malignant diseases; (3) transplants performed by EBMT centers; and (4) availability of clinical data at EBMT-Eurocord databases. Exclusion criteria were: (1) recipients of 2 or more CBUs; (2) recipients of CBU combined with another source of HSCs; (3) CBT with intrabone infusion; and/or (4) CBT with expanded in vitro CBU or any other form of experimental manipulations.
From January 1994 to December 2010, CBU samples were available for all 696 CBTs, and recipient samples were available for only 143 of them. Samples were shipped in dry ice to the Laboratory of Hematology (Ribeirão Preto School of Medicine, São Paulo University) according to international and Brazilian regulations for shipment of biological material. Informed consent procedures for performing genetic studies in CBUs and patients followed the ethical committee rules of each cord blood bank (CBB). Informed consents for CBU collection and CBT were obtained in accordance with Declaration of Helsinki. This study was approved by the local ethical committee (Comité de Protection des Personnes Ile-de-France IV) located in Saint-Louis Hospital, Paris, France.
Conditioning protocols, GVHD prophylaxis, selection of CBU, use of granulocyte-colony-stimulating factor, reactivation of cytomegalovirus (CMV) surveillance, and use of antimicrobial agents followed guidelines and rules of each transplant center.
Genetic polymorphism
Biological samples were shipped as extracted DNA, umbilical cord blood sample, or cord fragment tissue. Genomic DNA was extracted with the FlexiGene DNA kit (Qiagen) for CBU samples and the Wizard DNA Purification kit (Promega) for umbilical fragment tissue and stored in 1.5-mL Eppendorf tubes at a temperature of −20°C. DNA samples were genotyped by real-time polymerase chain reaction (PCR) assay for the following candidate genes related to immune response: NLRP1 (rs-5862), NLRP2 (rs-043684), NLRP3 (rs-10754558), TIRAP/Mal (rs-8177374), IL10 (rs-1800872), REL (rs-13031237), TNFRSF1B (rs-1061622), and CTLA-4 (rs-3087243). A complete list of all single-nucleotide polymorphisms (SNPs) studied and probes used for genotyping samples are described in supplemental Table 1 (available on the Blood Web site). Real-time PCR was performed by allelic discrimination method in the 7300 Real-Time PCR system, using TaqMan SNP genotyping assays and TaqMan genotyping Master Mix reagent (Applied Biosystems). Graphical interpretation of results was processed and supplied by ABI 7500 System SDS software (Applied Biosystems).
Statistical analysis
The primary objective of the study NRM was defined as death not related to recurrence of primary disease. Secondary outcomes were defined as follows: (1) overall survival (OS): time interval between transplantation and death due to any cause; (2) disease-free survival (DFS): time of life without relapse of the primary disease; (3) acute and chronic GVHD: diagnosis and grading were assigned by the transplant center using standard criteria38,39 ; (4) relapse of disease: event characterized by recurrence of the primary disease; (5) neutrophil and platelet engraftment: neutrophil count >0.5 × 109/L for 3 days and platelets >20 × 109/L for 3 days with no prior transfusion for at least 7 days.40,41
Disease status was classified according to criteria from the Center for International Blood and Marrow Transplant Research.42 Myeloablative conditioning was defined as a regimen containing either total body irradiation with a dose of >6 Gy, a dose of oral busulfan of >8 mg/kg, a dose of IV busulfan of >6.4 mg/kg, or a dose of treosulfan of at least 12 g/m2. Recipients and CBU were typed for HLA-A and -B at the antigenic level and for -DRB1 at the allelic level. A subset analysis using a homogenous group of patients with acute leukemia, given a myeloablative conditioning regimen and with available allele typing of HLA -A, -B, -C, and -DRB1, was performed (this subset of cohort 1 was defined as cohort 2). This cohort of patients was part of a previous study on the impact of HLA high-resolution typing on CBT outcomes, which has included a total of 1568 single CBTs.5
Preliminary analyses of Hardy-Weinberg equilibrium and minimum allele frequency of genotype distribution were performed. The χ2 test was used for measuring difference between groups.43,44 Independent risk factors for DFS and OS were performed by univariate and multivariate survival analyses with log-rank and Cox proportional hazards tests, respectively.45 Prognostic factors for neutrophil and platelet engraftment, acute and chronic GVHD, NRM were analyzed in a competitive risk scenario using Fine and Gray hazards proportional models, death being a competitive event.46 Multivariate models were constructed using variables that reached a P < .20 in univariate analysis and other variables with clinical relevance. Estimated type I error was set at 0.05. Statistical analyses were performed with SPSS 18.0 (SPSS Inc), Splus2000 (MathSoft), and The R Project for Statistical Computing (http://www.r-project.org).47,48
Results
Recipients, donors, and transplant characteristics
Patients, disease, donor (CBU), and transplant characteristics of the 3 cohorts of CBT recipients are listed in Table 1. The first cohort includes 696 patients who met the eligibility criteria of the study. The second cohort is a subset group of patients that includes 305 CBT recipients for whom HLA high-resolution typing of HLA -A, -B, -C, and -DRB1 of patients and CBUs were available. This cohort was used to confirm the results found in cohort 1. Supplemental Table 2 lists the 7 CBBs that provided the CBU samples for genotyping.
Recipients, donors, and CBT characteristics with available CBU samples and according to CTLA4-CBU genotyping (n = 696)
| Recipients . | AA-CBU genotyping, n = 162 . | AG-CBU genotyping, n = 349 . | GG-CBU genotyping, n = 185 . |
|---|---|---|---|
| Male sex, n | 79 | 200 | 110 |
| Age, median (range), y | 17 (0.4-64) | 16 (0.3-67) | 16 (0.6-69) |
| Weight, median (range), kg | 49 (5.5-95) | 50 (6-112) | 52 (6-119) |
| Children ≤18 y, n | 85 | 188 | 102 |
| Positive CMV serology, n | 56 | 125 | 76 |
| Major ABO incompatibility, n | 31 | 99 | 60 |
| Diagnostic, n | |||
| ALL | 63 | 135 | 76 |
| AML | 53 | 123 | 54 |
| MDS | 18 | 32 | 22 |
| CML | 9 | 16 | 9 |
| CLL | 3 | 2 | 0 |
| Lymphoma | 9 | 29 | 17 |
| Myeloma | 0 | 5 | 2 |
| Histiocytosis | 6 | 7 | 5 |
| Others | 1 | 0 | 0 |
| HLA compatibility, n | |||
| 6/6 | 19 | 35 | 14 |
| 5/6 | 59 | 136 | 71 |
| 4/6 | 75 | 155 | 88 |
| 3/6 | 3 | 13 | 6 |
| 2/6 | 2 | 1 | 3 |
| Disease status at time of CBT, n | |||
| Early | 56 | 119 | 50 |
| Intermediate | 58 | 129 | 73 |
| Advanced | 162 | 346 | 182 |
| Conditioning, n | |||
| Myeloablative | 130 | 283 | 143 |
| Nonmyeloablative | 32 | 62 | 38 |
| GVHD prophylaxis, n | |||
| CsA ± others | 143 | 295 | 158 |
| MTX ± others | 10 | 10 | 10 |
| Others | 2 | 7 | 4 |
| Use of ATG and /or monoclonal antibody, n | |||
| Yes | 121 | 281 | 136 |
| No | 34 | 52 | 38 |
| Infused TNC dose, median (range), ×107/kg | 3.5 (0.9-30) | 3.3 (0.6-22) | 3.4 (0.7-21) |
| Infused CD34+ cell dose, median (range), ×105/kg | 1.5 (0.2-20) | 1.5 (0.2-39) | 1.4 (0.3-18) |
| Follow-up (range), mo | 40 (4-181) | 55 (1-160) | 48 (1-195) |
| Recipients . | AA-CBU genotyping, n = 162 . | AG-CBU genotyping, n = 349 . | GG-CBU genotyping, n = 185 . |
|---|---|---|---|
| Male sex, n | 79 | 200 | 110 |
| Age, median (range), y | 17 (0.4-64) | 16 (0.3-67) | 16 (0.6-69) |
| Weight, median (range), kg | 49 (5.5-95) | 50 (6-112) | 52 (6-119) |
| Children ≤18 y, n | 85 | 188 | 102 |
| Positive CMV serology, n | 56 | 125 | 76 |
| Major ABO incompatibility, n | 31 | 99 | 60 |
| Diagnostic, n | |||
| ALL | 63 | 135 | 76 |
| AML | 53 | 123 | 54 |
| MDS | 18 | 32 | 22 |
| CML | 9 | 16 | 9 |
| CLL | 3 | 2 | 0 |
| Lymphoma | 9 | 29 | 17 |
| Myeloma | 0 | 5 | 2 |
| Histiocytosis | 6 | 7 | 5 |
| Others | 1 | 0 | 0 |
| HLA compatibility, n | |||
| 6/6 | 19 | 35 | 14 |
| 5/6 | 59 | 136 | 71 |
| 4/6 | 75 | 155 | 88 |
| 3/6 | 3 | 13 | 6 |
| 2/6 | 2 | 1 | 3 |
| Disease status at time of CBT, n | |||
| Early | 56 | 119 | 50 |
| Intermediate | 58 | 129 | 73 |
| Advanced | 162 | 346 | 182 |
| Conditioning, n | |||
| Myeloablative | 130 | 283 | 143 |
| Nonmyeloablative | 32 | 62 | 38 |
| GVHD prophylaxis, n | |||
| CsA ± others | 143 | 295 | 158 |
| MTX ± others | 10 | 10 | 10 |
| Others | 2 | 7 | 4 |
| Use of ATG and /or monoclonal antibody, n | |||
| Yes | 121 | 281 | 136 |
| No | 34 | 52 | 38 |
| Infused TNC dose, median (range), ×107/kg | 3.5 (0.9-30) | 3.3 (0.6-22) | 3.4 (0.7-21) |
| Infused CD34+ cell dose, median (range), ×105/kg | 1.5 (0.2-20) | 1.5 (0.2-39) | 1.4 (0.3-18) |
| Follow-up (range), mo | 40 (4-181) | 55 (1-160) | 48 (1-195) |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; CsA, cyclosporin A; MDS, myelodysplastic syndrome; MTX, methotrexate.
Of the first cohort, 56% of patients were male (n = 380), 54% children (n = 375), 61% had a positive CMV serology (n = 393), and 34% received a CBT with a major ABO incompatibility (n = 190). Ten percent of CBT were HLA-identical (6 of 6) (n = 68), 39% were transplanted with 1 HLA disparity (5 of 6) (n = 266), 51% were transplanted with 2 or more HLA disparities (4 of 6 or 3 of 6) (n = 346). At time of transplantation, recipients had early disease status in 33% (n = 225), intermediate disease in 38% (n = 260), and advanced disease in 27% (n = 205) of the cases. GVHD prophylaxis was cyclosporine-based in 91% of patients (n = 597). Myeloablative conditioning was used in 80% (n = 556) and antithymocyte immunoglobulin (ATG) or monoclonal antibody in 81% (n = 538) of the cases. Infused median total nucleated cells (TNCs) were 3.4 × 107/kg (range, 0.6-30) and median CD34+ cells were 1.5 × 105/kg (range, 0.2-39). Median follow-up was 49 (range, 0.8-195) months. Table 1 describes recipients, donors, and CBT characteristics of cohort 1 according to CTLA4-CBU genotyping.
In the second cohort, all patients had acute leukemia and received a myeloablative conditioning regimen, 66% were children (n = 191), and 91% received ATG (n = 269). Only 5% of patients were transplanted with an 8 of 8 CBU graft (n = 15), whereas 13% were 7 of 8 (n = 40), 25% were 6 of 8 (n = 76), 32% were 5 of 8 (n = 98), 19% were 4 of 8 (n = 59), and 6% were 3 of 8 (n = 17). Table 2 describes recipients, donors, and CBT characteristics of cohort 2 according to CTLA4-CBU genotyping.
Recipients, donors, and CBT characteristics with available CBU samples, HLA-HR data, and according to CTLA4-CBU genotyping (n = 305)
| Recipients . | AA-CBU genotyping, n = 73 . | AG-CBU genotyping, n = 153 . | GG-CBU genotyping, n = 79 . |
|---|---|---|---|
| Male sex, n | 32 | 93 | 51 |
| Age, median (range), y | 13 (0.7-56) | 13 (0.8-60) | 11 (0.7-48) |
| Weight, median (range), kg | 39 (7-93) | 50 (7-112) | 36 (6-94) |
| Children ≤18 y, n | 45 | 91 | 55 |
| Positive CMV serology, n | 43 | 91 | 46 |
| Major ABO incompatibility, n | 13 | 49 | 29 |
| Diagnostic, n | |||
| ALL | 36 | 82 | 42 |
| AML | 28 | 57 | 24 |
| MDS | 9 | 14 | 13 |
| HLA compatibility, n | |||
| 8/8 | 5 | 8 | 2 |
| 7/8 | 9 | 17 | 14 |
| 6/8 | 18 | 40 | 18 |
| 5/8 | 21 | 45 | 32 |
| 4/8 | 13 | 34 | 12 |
| 3/8 | 7 | 9 | 1 |
| Disease status at time of CBT, n | |||
| Early | 32 | 66 | 27 |
| Intermediate | 28 | 56 | 30 |
| Advanced | 13 | 31 | 22 |
| GVHD prophylaxis, n | |||
| CsA ± others | 68 | 137 | 71 |
| MTX ± others | 4 | 9 | 3 |
| Others | 0 | 2 | 3 |
| Use of ATG and /or monoclonal antibody, n | |||
| Yes | 8 | 9 | 9 |
| No | 62 | 139 | 68 |
| Infused TNC dose, median (range), ×107/kg | 3.7 (1-17) | 3.4 (1-19) | 3.9 (1-20) |
| Infused CD34+ cell dose, median (range), ×105/kg | 1.8 (0.3-18) | 1.7 (0.2-9) | 1.6 (0.3-17) |
| Follow-up (range), mo | 59 (4-125) | 60 (3-145) | 40 (6-86) |
| Recipients . | AA-CBU genotyping, n = 73 . | AG-CBU genotyping, n = 153 . | GG-CBU genotyping, n = 79 . |
|---|---|---|---|
| Male sex, n | 32 | 93 | 51 |
| Age, median (range), y | 13 (0.7-56) | 13 (0.8-60) | 11 (0.7-48) |
| Weight, median (range), kg | 39 (7-93) | 50 (7-112) | 36 (6-94) |
| Children ≤18 y, n | 45 | 91 | 55 |
| Positive CMV serology, n | 43 | 91 | 46 |
| Major ABO incompatibility, n | 13 | 49 | 29 |
| Diagnostic, n | |||
| ALL | 36 | 82 | 42 |
| AML | 28 | 57 | 24 |
| MDS | 9 | 14 | 13 |
| HLA compatibility, n | |||
| 8/8 | 5 | 8 | 2 |
| 7/8 | 9 | 17 | 14 |
| 6/8 | 18 | 40 | 18 |
| 5/8 | 21 | 45 | 32 |
| 4/8 | 13 | 34 | 12 |
| 3/8 | 7 | 9 | 1 |
| Disease status at time of CBT, n | |||
| Early | 32 | 66 | 27 |
| Intermediate | 28 | 56 | 30 |
| Advanced | 13 | 31 | 22 |
| GVHD prophylaxis, n | |||
| CsA ± others | 68 | 137 | 71 |
| MTX ± others | 4 | 9 | 3 |
| Others | 0 | 2 | 3 |
| Use of ATG and /or monoclonal antibody, n | |||
| Yes | 8 | 9 | 9 |
| No | 62 | 139 | 68 |
| Infused TNC dose, median (range), ×107/kg | 3.7 (1-17) | 3.4 (1-19) | 3.9 (1-20) |
| Infused CD34+ cell dose, median (range), ×105/kg | 1.8 (0.3-18) | 1.7 (0.2-9) | 1.6 (0.3-17) |
| Follow-up (range), mo | 59 (4-125) | 60 (3-145) | 40 (6-86) |
This cohort is a subset of recipients with available CBU (n = 696).
HR-HLA, high-resolution HLA. Other abbreviations are explained in Table 1.
Genetic polymorphism
Results of CBU and recipient gene polymorphisms and their prevalence are listed in supplemental Tables 3 and 4. Alleles or genotypes with <1% of frequency were not observed and frequencies were similar among groups. In addition, group allelic frequencies were in Hardy-Weinberg equilibrium.
Outcomes
Neutrophil and platelet engraftment.
Cumulative incidence of neutrophil recovery was 82% (95% confidence interval [CI], 79%-85%) at day 60 post-CBT. According to CBU genotypes, univariate analysis showed an association of CTLA4-CBU and neutrophil recovery. In fact, at day 60, cumulative incidence of neutrophil recovery for patients transplanted with an AA-CTLA4-CBU was 85% (95% CI, 79%-90%), whereas it was 84% (95% CI, 80%-87%) for AG, and 77% (95% CI, 70%-83%) for GG genotypes, respectively. Multivariate analysis confirmed a delayed neutrophil recovery for recipients of CBU with GG-CTLA4 genotype (hazard ratio [HR], 1.33; 95% CI, 1.04-1.70; P = .02). Others factors independently associated with neutrophil recovery were: age >18 years (HR, 1.58; 95% CI, 1.28-1.96; P < .01), advanced status of disease at time of CBT (HR, 1.27; 95% CI, 1.03-1.54; P = .02), and median infused TNC (>median; HR, 0.71; 95% CI, 0.66-0.93; P = .01). Details of multivariate analysis for neutrophil engraftment are shown in supplemental Tables 5 and 6.
At day 180, cumulative incidence of platelet recovery was 57% (95% CI, 53%-61%). In the multivariate model, platelet recovery independently associated with: age >18 years (HR, 1.38; 95% CI, 1.06-1.79; P = .02), advanced disease at time of CBT (HR, 1.32; 95% CI, 1.02-1.69; P = .03), and recipients with negative CMV serology status (HR, 0.77; 95% CI, 0.62-0.94; P = .01). Details of multivariate analysis for platelet engraftment are shown in supplemental Tables 7 and 8.
Acute and chronic GVHD.
Cumulative incidence of grade II-IV acute GVHD was 31% (95% CI, 28%-35%) at day 100. No statistical association was found between any SNP analyzed and the incidence of acute GVHD. In multivariate analysis, variables associated with acute GVHD were: age >18 years (HR, 1.69; 95% CI, 1.51-1.92; P < .01), myeloablative conditioning (HR, 1.77; 95% CI, 1.57-2.01; P < .01), use of ATG or monoclonal antibody (HR, 0.57; 95% CI, 0.50-0.64; P < .01), and advanced disease at CBT (HR, 1.39; 95% CI, 1.23-1.57; P = .03). At 4 years, cumulative incidence of chronic GVHD was 16% (95% CI, 13%-19%). No statistical association between the candidate genes and chronic GVHD was observed. In multivariate analysis, only advanced disease at time of CBT was associated with a higher incidence of chronic GVHD (HR, 1.74; 95% CI, 1.54-1.96; P = .03). Details of multivariate analysis for acute or chronic GVHD are shown in supplemental Tables 9-12.
NRM, OS, and causes of death.
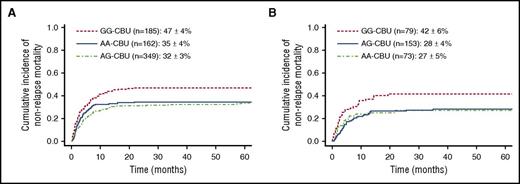
Cumulative incidence of NRM was 37% (95% CI, 33%-41%) at 4 years. Univariate analysis according to CBU genotype showed higher NRM for recipients of GG-CTLA4-CBU genotype (AA, 35% [95% CI, 29%-43%]; AG, 32% [95% CI, 27%-37%]; GG, 47% [95% CI, 40%-54%]) (Figure 1A). Multivariate analysis confirmed these results demonstrating higher NRM for recipients of GG-CBU genotype (HR, 1.50; 95% CI, 1.11-2.03; P < .01). Other factors independently associated with NRM were: previous HCT (HR, 1.73; 95% CI, 1.20-2.49; P < .01), recipients with negative CMV serology status (HR, 0.63; 95% CI, 0.47-0.85; P < .01), use of ATG or monoclonal antibody (HR, 1.69; 95% CI, 1.07-2.67; P = .03), and advanced disease at time of CBT (HR, 1.72; 95% CI, 1.30-2.27; P < .01). In the analysis for cohort 2, recipients of CBU GG CTLA4 genotype had increased NRM (HR, 1.85; 95% CI 1.14-3.00; P = .01). Four-year cumulative incidence of NRM was 27% (95% CI, 18%-38%) for AA, 28% (95% CI, 21%-36%) for AG, and 42% (95% CI, 31%-54%) for GG (Figure 1B). Details of multivariate analysis for NRM are shown in supplemental Tables 13 and 14.
NRM according to CBUgenotype. (A) CTLA4 for recipients with available CBU samples (n = 696). (B) CTLA4 for recipients with available CBU samples and HLA high-resolution typing (n = 305).
NRM according to CBUgenotype. (A) CTLA4 for recipients with available CBU samples (n = 696). (B) CTLA4 for recipients with available CBU samples and HLA high-resolution typing (n = 305).
Estimated OS at 4 years was 40% (95% CI, 36%-44%). Univariate analysis by CBU genotype showed inferior OS for recipients of GG-CTLA4-CBU genotype (AA, 47% [95% CI, 39%-55%]; AG, 41% [95% CI, 35%-47%]; GG, 33% [95% CI, 26%-42%]), and multivariate analysis confirmed these results (HR, 1.41; 95% CI, 1.04-1.90; P = .02). Other factors independently associated with OS were: negative CMV serology status (HR, 0.69; 95% CI, 0.55-0.87; P < .01), intermediate or advanced disease at time of CBT (HR, 1.50; 95% CI, 1.13-1.94; P < .01 and HR, 2.20; 95% CI, 1.67-2.89; P < .01, respectively), and >2 HLA incompatibility (HR, 1.26; 95% CI, 1.01-1.58; P = .04). In the analysis of cohort 2, recipients of GG-CTLA4-CBU genotype had inferior OS, but with borderline statistical significance (HR, 1.54; 95% CI, 1.03-2.41; P = .06). At 4 years, OS was 53% (95% CI, 41%-64%) for AA, 45% (95% CI, 37%-53%) for AG, and 33 (95% CI, 23%-46%) for GG. Details of multivariate analysis for OS are shown in supplemental Tables 15 and 16.
Fifty-nine percent of recipients died (n = 412). Sixty-two percent of deaths were related-transplant complications (n = 255), 36% to relapse or disease progression (n = 149), whereas 2% were of unknown causes (n = 8). Deaths related to transplant complications (n = 255) were due to: infections, 44% (n = 113); GVHD, 23% (n = 59); multiple organ failure, 6% (n = 16); pulmonary complications, 6% (n = 13); graft failure, 5% (n = 12); bleeding disorders, 5% (n = 12); hepatic veno-occlusive disease, 4% (n = 11); and other or unknown causes, 7% (n = 19) . Associations between SNPs and causes of death were studied by correspondence analysis. Recipients of CBU with GG-CTLA4 genotype died mainly of transplant complications, in particular, GVHD and infectious complications (Table 3).
Cause of deaths of recipients according to CTLA4-CBU genotyping
| Variable . | Recipients with available CBU samples, N = 696 . | ||
|---|---|---|---|
| AA, n = 162 . | AG, n = 349 . | GG, n = 185 . | |
| GVHD | 13 | 24 | 22 |
| Infections | 22 | 55 | 36 |
| Pulmonary complications | 3 | 5 | 5 |
| Bleeding disorders | 2 | 6 | 4 |
| Multiple organ failure | 6 | 4 | 6 |
| Graft failure | 3 | 3 | 6 |
| VOD | 4 | 4 | 3 |
| Others | 3 | 14 | 2 |
| Total | 56 | 115 | 84 |
| Variable . | Recipients with available CBU samples, N = 696 . | ||
|---|---|---|---|
| AA, n = 162 . | AG, n = 349 . | GG, n = 185 . | |
| GVHD | 13 | 24 | 22 |
| Infections | 22 | 55 | 36 |
| Pulmonary complications | 3 | 5 | 5 |
| Bleeding disorders | 2 | 6 | 4 |
| Multiple organ failure | 6 | 4 | 6 |
| Graft failure | 3 | 3 | 6 |
| VOD | 4 | 4 | 3 |
| Others | 3 | 14 | 2 |
| Total | 56 | 115 | 84 |
VOD, hepatic veno-oclusive disease.
Relapse.
Cumulative incidence of relapse was 28% (95% CI, 24%-32%) at 4 years. In univariate analysis, CTLA4 genotype was associated with higher incidence of relapse. It was 22% (95% CI, 17%-28%) for AA, 33% (95% CI, 29%-37%) for AG, and 25% (95% CI, 20%-31%) for GG. However, multivariate analysis did not confirm this result. Other variables independently associated with relapse were: use of ATG or monoclonal antibody (HR, 1.75; 95% CI, 1.15-2.70; P < .01) and advanced disease at time of CBT (HR, 1.83; 95% CI, 1.32-2.56; P < .01). In cohort 2, CTLA4-CBU genotype was not associated with relapse. Details of multivariate analysis for NRM are shown in supplemental Tables 17 and 18.
Disease-free survival.
At 4 years, DFS was 35% (95% CI, 31%-39%). According to CBU genotype, univariate analysis demonstrated an impact for CTLA4 on DFS (AA, 43% [95% CI, 35%-51%]; AG, 35% [95% CI, 29%-41%]; and GG, 29% [95% CI, 24%-35%]). Multivariate analysis confirmed these results showing inferior DFS of recipients receiving GG genotype CBU (HR, 1.41; 95% CI, 1.06-1.88; P = .02). Other variables associated with DFS were: recipients with negative CMV serology status (HR, 0.74; 95% CI, 0.60-0.92; P = .01), intermediate disease at time of CBT (HR, 1.46; 95% CI, 1.13-1.89; P < .01), and advanced disease at time of CBT (HR, 2.17; 95% CI, 1.67-2.82; P < .01). In cohort 2, recipients of GG-CTLA4-CBU genotype had inferior DFS, but it was not statically significant (HR, 1.49; 95% CI, 0.96-2.30; P = .07). Details of multivariate analysis for NRM are shown in supplemental Tables 19 and 20.
Table 4 shows a summary of the multivariate analysis results according to CBU genotype.
Summary of multivariate analysis results according to CBU genotype
| Analyzed cohort . | Outcomes . | SNP . | HR . | 95% CI . | P . | |
|---|---|---|---|---|---|---|
| < . | > . | |||||
| Recipients with available CBU samples, n = 696 | Neutrophil engraftment | CTLA4 genotype AA | Reference group | .08 | ||
| CTLA4 genotype AG | 0.85 | 0.69 | 1.05 | .14 | ||
| CTLA4 genotype GG | 1.33 | 1.04 | 1.70 | .02 | ||
| NRM | CTLA4 genotype AA | Reference group | .02 | |||
| CTLA4 genotype AG | 0.68 | 0.47 | 0.98 | .04 | ||
| CTLA4 genotype GG | 1.50 | 1.11 | 2.03 | <.01 | ||
| OS | CTLA4 genotype AA | Reference group | .09 | |||
| CTLA4 genotype AG | 1.19 | 0.91 | 1.56 | .21 | ||
| CTLA4 genotype GG | 1.41 | 1.04 | 1.90 | .02 | ||
| Relapse | CTLA4 genotype AA | Reference group | .07 | |||
| CTLA4 genotype AG | 1.35 | 0.83 | 2.17 | .22 | ||
| CTLA4 genotype GG | 1.19 | 0.82 | 1.27 | .35 | ||
| DFS | CTLA4 genotype AA | Reference group | .06 | |||
| CTLA4 genotype AG | 1.21 | 0.93 | 1.57 | .15 | ||
| CTLA4 genotype GG | 1.41 | 1.06 | 1.88 | .02 | ||
| Recipients with available CBU samples and HR-HLA data (n = 305)* | Neutrophil engraftment | CTLA4 genotype AA | Reference group | .16 | ||
| CTLA4 genotype AG | 0.74 | 0.54 | 1.01 | .07 | ||
| CTLA4 genotype GG | 1.33 | 0.92 | 1.92 | .13 | ||
| NRM | CTLA4 genotype AA | Reference group | .05 | |||
| CTLA4 genotype AG | 0.68 | 0.38 | 1.2 | .18 | ||
| CTLA4 genotype GG | 1.85 | 1.14 | 3.00 | .01 | ||
| OS | CTLA4 genotype AA | Reference group | .07 | |||
| CTLA4 genotype AG | 1.05 | 0.70 | 1.59 | .81 | ||
| CTLA4 genotype GG | 1.54 | 0.99 | 2.41 | .06 | ||
| Relapse | CTLA4 genotype AA | Reference group | .25 | |||
| CTLA4 genotype AG | 1.49 | 0.75 | 2.94 | .66 | ||
| CTLA4 genotype GG | 1.12 | 0.65 | 1.96 | .25 | ||
| DFS | CTLA4 genotype AA | Reference group | .17 | |||
| CTLA4 genotype AG | 1.16 | 0.78 | 1.73 | .46 | ||
| CTLA4 genotype GG | 1.49 | 0.96 | 2.30 | .07 | ||
| Analyzed cohort . | Outcomes . | SNP . | HR . | 95% CI . | P . | |
|---|---|---|---|---|---|---|
| < . | > . | |||||
| Recipients with available CBU samples, n = 696 | Neutrophil engraftment | CTLA4 genotype AA | Reference group | .08 | ||
| CTLA4 genotype AG | 0.85 | 0.69 | 1.05 | .14 | ||
| CTLA4 genotype GG | 1.33 | 1.04 | 1.70 | .02 | ||
| NRM | CTLA4 genotype AA | Reference group | .02 | |||
| CTLA4 genotype AG | 0.68 | 0.47 | 0.98 | .04 | ||
| CTLA4 genotype GG | 1.50 | 1.11 | 2.03 | <.01 | ||
| OS | CTLA4 genotype AA | Reference group | .09 | |||
| CTLA4 genotype AG | 1.19 | 0.91 | 1.56 | .21 | ||
| CTLA4 genotype GG | 1.41 | 1.04 | 1.90 | .02 | ||
| Relapse | CTLA4 genotype AA | Reference group | .07 | |||
| CTLA4 genotype AG | 1.35 | 0.83 | 2.17 | .22 | ||
| CTLA4 genotype GG | 1.19 | 0.82 | 1.27 | .35 | ||
| DFS | CTLA4 genotype AA | Reference group | .06 | |||
| CTLA4 genotype AG | 1.21 | 0.93 | 1.57 | .15 | ||
| CTLA4 genotype GG | 1.41 | 1.06 | 1.88 | .02 | ||
| Recipients with available CBU samples and HR-HLA data (n = 305)* | Neutrophil engraftment | CTLA4 genotype AA | Reference group | .16 | ||
| CTLA4 genotype AG | 0.74 | 0.54 | 1.01 | .07 | ||
| CTLA4 genotype GG | 1.33 | 0.92 | 1.92 | .13 | ||
| NRM | CTLA4 genotype AA | Reference group | .05 | |||
| CTLA4 genotype AG | 0.68 | 0.38 | 1.2 | .18 | ||
| CTLA4 genotype GG | 1.85 | 1.14 | 3.00 | .01 | ||
| OS | CTLA4 genotype AA | Reference group | .07 | |||
| CTLA4 genotype AG | 1.05 | 0.70 | 1.59 | .81 | ||
| CTLA4 genotype GG | 1.54 | 0.99 | 2.41 | .06 | ||
| Relapse | CTLA4 genotype AA | Reference group | .25 | |||
| CTLA4 genotype AG | 1.49 | 0.75 | 2.94 | .66 | ||
| CTLA4 genotype GG | 1.12 | 0.65 | 1.96 | .25 | ||
| DFS | CTLA4 genotype AA | Reference group | .17 | |||
| CTLA4 genotype AG | 1.16 | 0.78 | 1.73 | .46 | ||
| CTLA4 genotype GG | 1.49 | 0.96 | 2.30 | .07 | ||
Abbreviations are explained in Table 2.
This cohort is a subset of recipients with available CBU (n = 696).
Discussion
CBT is a valuable option for patients with malignant diseases lacking an HLA-identical donor. Since the first CBT in 1988, considerable progress has been made in this field.49 As in allogeneic HSCT (allo-HSCT), many variables, mostly related to CBU, recipients, disease, or transplantation characteristics, influence the occurrence of CBT complications, such as transplantation toxicities, GVHD, infections, and relapse. In addition, CBT outcomes are comparable to other HSC sources when CBT is performed with <2 of 6 HLA mismatches and adequate cell dose.50-54 However, the influence of non-HLA genetic factors, such as genetic polymorphism of immune response, on CBT outcomes has not been well investigated. The purpose of the current study was to investigate the association between CBU and recipient genotypes with CBT outcomes. With this aim, CBUs (n = 696) were genotyped for 8 SNPs related to immunological response. Before establishing any association between genotypes and phenotypes, preliminary analysis ruled out genotyping errors by minimum allelic frequency test (data not shown).
In this analysis, polymorphisms of innate immune response were not associated with CBT outcomes. CTLA4 was the only candidate gene of adaptive immune response evaluated in this study. Interestingly, recipients of CBU carrying GG-CTLA4 genotype had lower neutrophil recovery, increased NRM, and decreased DFS. In addition, recipients of CBU carrying the AA-CTLA4 genotype showed a lower incidence of relapse. Furthermore, increased NRM was confirmed in recipients of CBU with GG-CTLA4 with acute leukemia, receiving myeloablative conditioning, and with available high-resolution HLA typing.
CTLA4 and its genetic variations have been largely investigated in the field of allo-HSCT, especially regarding GVHD and survival. However, because of contradictory results, the real impact of CTLA4 on outcomes is difficult to evaluate.28-33 The largest study available, with a homogeneous cohort, yielded negative results. Nevertheless, there is no report in the literature describing the role of CTLA4 in CBT.13
The impact of CTLA4 presented in our study is biologically plausible. It has been suggested that the G allele of CT60 produces lower messenger RNA of soluble CTLA4 (sCTLA4) whose expression is the functional basis for the observed association between autoimmune diseases and CTLA4.55 Others have analyzed messenger RNA expression of sCTLA4 in 60 healthy blood donors and have identified allele A as being responsible for greater production of sCTLA4. This study also reported that allo-HSCT recipients of progenitor cells from AA genotype donors had higher number of alloimmune reactions, as indicated by the higher incidence of acute GVHD. Furthermore, the G allele conferred increased risk of disease relapse.32 Moreover, it has been suggested that sCTLA4 inhibits the B7-flCTLA4 complex, with consequent influence on T-cell alloreactivity.32,55 These previous findings, taken together, are in agreement with the results observed in our study.
In the current study, CBU with GG genotype was associated with decreased neutrophil engraftment, increased NRM, and decreased survival rates. Higher cell dose and fewer HLA disparities of the CBU graft with the recipient are known factors enhancing engraftment after CBT.1 The number of hematopoietic progenitor cells and lymphocytes in CBUs is, usually, 1 log less than in other HSC grafts, and both of these types of cells have an important role in engraftment after allo-HSCT. Therefore, we could speculate that lymphocytes carrying the GG-CTLA4 genotype of CBU have lower alloreactivity, which could, in turn, explain the impact of CTLA4 on engraftment. Analyzing CBT immune tolerance, Miller et al compared CTLA4 gene expression of CBU and HSCs of adult donors and demonstrated reduced expression of CTLA4 in CBU T cells, but theses results were not reproduced in accordance with CTLA4 allele genotype.56 Another interesting finding in our study was that recipients of CBU AA genotype experienced lower relapse. Again, this result suggests superior T-cell response for AA-CBU genotype.
Although our findings are based on solid research and are biologically likely, there are some limitations to our study. First, this has a retrospective registry-based nature, with multiple transplant centers and CBBs. Second, the effect of recipient CTLA4 genotype could not be studied as recipient samples were not available. Finally, the study population is heterogeneous, with patients with different diseases receiving different types of conditioning regimen and transplanted in a wide range period. To overcome the limitations, the final multivariate models were adjusted for these variables, however, cautions on the conclusions should be taken when performing multiple tests. Therefore, with the aim of circumventing these limitations, we performed the same analysis in a homogeneous subset of cohort 1 (cohort 2) to have a more homogeneous population, with only patients transplanted for acute leukemia all given a myeloablative conditioning regimen and with available allele HLA typing. In this cohort, we were able to demonstrate that CTLA4 was associated with NRM, but not with other outcomes. The absence of association with other outcomes may have been related to the limited sample size. Nevertheless, caution should be taken when interpreting the conclusions, as when performing multiple tests, some of the results may have arisen by chance. In addition, studies on functional analysis of the CTLA4 gene and the impact of HLA-DP on CBT outcomes are also needed.57
In many retrospective studies, the number of HLA disparities and cell dose are important factors associated with outcomes after CBT and are frequently used to select the best CBUs.1,53 However, when multiple CBUs are available for the same patient, CTLA4-CBU genotype could be tested and used as an additional criterion to select the CBU, with the aim of improving outcomes after CBT.
In conclusion, gene polymorphisms of the immune response may influence CBT outcomes. CTLA4 was shown to impact CBT outcomes according to CBU genotype. The association of GG-CTLA4 of the CBU with lower survival and higher NRM suggests that this polymorphism might be considered for CBU selection when >1 CBU meeting the current suggested selection criteria of cell dose and HLA matching is available. Importantly, CTLA4 typing of CBUs should not, significantly, increase costs or delay transplantation, and it could be provided by the CBBs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all participating cord blood banks and transplant centers. The authors also thank Dalva Tereza Catto for valuable contribution on samples shipment from Europe to the Laboratory of Hematology of Ribeirão Preto School of Medicine of São Paulo University.
V.R. was supported by National Health Service Blood and Transplants and funded by the National Institute for Health Research Biomedical Research Centers funding scheme, Oxford, United Kingdom, and by Fundação de Amparo à Pesquisa do Estado de São Paulo grant 2013/02162-8, São Paulo, Brazil. This work was supported by the Center for Cell-Based Therapy of Ribeirão Preto School of Medicine of São Paulo University.
Authorship
Contribution: R.C., M.A.Z., E.G., and V.R. designed research; R.C., R.A.P., and M.T. performed experiments; R.C., F.V., and A.R. verified clinical data; S.Q., F.P., G.K., J.L.V., P.B., R.S., C.H.L., and C.D.-d.-H. provided biological samples; G.S., G.M., and H.B. provided clinical data; R.C., F.F., J.P., and V.R. analyzed the data and performed statistical analysis; and R.C., E.G., and V.R. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Eurocord, Cord Blood Committee Cellular Therapy–Immunobiology Working Party of the European Society for Blood and Marrow Transplantation, Netcord and Faculdade de Medicina de Ribeirão Preto–Faculdade de Medicina de São Paulo, Universidade de São Paulo appears in “Appendix.”
Correspondence: Renato Cunha, Bone Marrow Transplantation Unit, Clinical Hospital of Ribeirão Preto School of Medicine of São Paulo University, São Paulo, Brazil; e-mail: renatolgc@gmail.com.
Appendix: study group members
The study group members are as follows: Eurocord: R.C., F.V., A.R., E.G., V.R.; Cord Blood Committee Cellular Therapy–Immunobiology Working Party of the European Society for Blood and Marrow Transplantation: A.R.; Netcord: S.Q., F.P., G.K., J.L.V., P.B., R.S., C.H.L.; Faculdade de Medicina de Ribeirão Preto–Faculdade de Medicina de São Paulo, Universidade de São Paulo: R.C., M.A.Z., M.T., R.A.P., V.R.