Key Points
Variants in GP1BB can cause autosomal dominant macrothrombocytopenia.
Abstract
The von Willebrand receptor complex, which is composed of the glycoproteins Ibα, Ibβ, GPV, and GPIX, plays an essential role in the earliest steps in hemostasis. During the last 4 decades, it has become apparent that loss of function of any 1 of 3 of the genes encoding these glycoproteins (namely, GP1BA, GP1BB, and GP9) leads to autosomal recessive macrothrombocytopenia complicated by bleeding. A small number of variants in GP1BA have been reported to cause a milder and dominant form of macrothrombocytopenia, but only 2 tentative reports exist of such a variant in GP1BB. By analyzing data from a collection of more than 1000 genome-sequenced patients with a rare bleeding and/or platelet disorder, we have identified a significant association between rare monoallelic variants in GP1BB and macrothrombocytopenia. To strengthen our findings, we sought further cases in 2 additional collections in the United Kingdom and Japan. Across 18 families exhibiting phenotypes consistent with autosomal dominant inheritance of macrothrombocytopenia, we report on 27 affected cases carrying 1 of 9 rare variants in GP1BB.
Introduction
The earliest step in hemostasis is the tethering of platelets to the damaged endothelium through their cell surface receptor for von Willebrand factor (VWF). The glycoproteins (GPs) Ibα and Ibβ, in association with GPV and GPIX, form the transmembrane receptor complex on platelets for VWF.1,2 The leucine-rich repeat (LRR) domain of GPIbα contains the binding site for VWF, whereas the β chain contributes to the surface expression of the complex, which, by phosphorylating its intracellular domain, also participates in downstream signaling. GPIbβ is synthesized from a 1.5-kb mRNA transcribed from GP1BB and is highly expressed in megakaryocytes, but absent from other blood cell progenitors.3 The mRNA encodes a 206–amino-acid-long transmembrane protein with a 22-kD molecular mass and an extracellular LRR domain.4-6
Bernard and Soulier were the first to describe a patient with a rare autosomal recessive syndrome (Bernard-Soulier syndrome; BSS) characterized by giant platelets with concomitant thrombocytopenia complicated by severe bleeding.7 Platelets from patients with this syndrome do not agglutinate in response to ristocetin and show a subtly reduced aggregation response to thrombin. Over the course of 4 decades, 45, 39, and 28 variants have been identified in GP1BA, GP1BB, and GP9, respectively, that, if present in compound heterozygous or homozygous form, cause the VWF receptor to be absent or reduced in function.8 Only 4 variants in GP1BA with dominant effects have been reported, one being the “Bolzano variant,”9 and a further 3 observed in 3 isolated pedigrees.8 Only 2 variants in GP1BB have been reported as being possibly responsible for dominant macrothrombocytopenia. One encoding Y113C was identified in 2 Japanese families and led to initial speculation that variants in the subunits of the GPIb/IX complex could span a spectrum of platelet and bleeding phenotypes.10,11 Another, encoding R42C, was observed in only 1 Japanese patient, and data from family or functional studies supporting an association with a dominant phenotype were lacking.12 There are no reports of variants in GP9 having a dominant effect, and BSS cases resulting from variants in GP5 do not exist because GPV is not required for receptor expression.13
We found a significant statistical association between rare monoallelic nonsynonymous variants in GP1BB and macrothrombocytopenia. Family history and cosegregation data from 18 pedigrees, in which 27 affected cases carry 1 of 9 rare variants in GP1BB, are consistent with autosomal dominant inheritance.
Study design
Study population
The study population comprises a discovery collection and 2 validation collections. The discovery collection consists of cases with an assumed inherited bleeding or platelet disorder (BPD) of unknown molecular etiology or their relatives enrolled in the National Institute for Health Research (NIHR) BioResource after providing informed written consent.14 The BioResource cohort was enrolled during a 4-year period and is composed of 1542 patients with a BPD or their close relatives, with a further 5422 patients with other rare inherited disorders or their close relatives (https://bioresource.nihr.ac.uk/rare-diseases/clinicians/). The validation collections contain data from 75 patients with thrombocytopenia assessed with the ThromboGenomics diagnostic platform,15 and 301 Japanese probands16 referred during an 8-year period for a suspected diagnosis of inherited macrothrombocytopenia and their relatives. Institutional review board approval was obtained for this international multicenter study.
Data analysis
Coding of clinical and laboratory phenotypes with Human Phenotype Ontology (HPO) terms and collection of numerical and family history data were performed as described previously.14 Variants from high-throughput sequencing were called and annotated using Isaac (Illumina, Inc; whole-genome sequencing) or as described previously15 (capture-based sequencing). We used phenotype similarity regression17 (SimReg) to identify statistical associations between presence of a variant affecting protein sequence in a gene and similarity to a latent HPO–coded phenotype in the discovery collection, subsequently corroborated using Fisher’s exact test. The variants obtained by high-throughput DNA sequencing of the probands were genotyped by Sanger sequencing in relatives who agreed to participate in this study. The variants identified in all members of the Japanese collection were obtained by Sanger sequencing. Where possible, complete blood counts were obtained using automated hematology analyzers, VWF receptor expression levels on platelets were determined by cytometry, and platelet function was tested by light transmission aggregometry. Macrothrombocytopenia was deemed present if the platelet count was below 150 × 109/L, the mean platelet volume was above 12 fL, or there was clear platelet anisocytosis, with a subset of platelets being abnormally large.
Results and discussion
We identified a strong statistical association between the presence of rare nonsynonymous monoallelic DNA variants (population allele frequency less than 1/10 000) in GP1BB and macrothrombocytopenia in the discovery collection (SimReg posterior probability = 0.93 with inferred characteristic phenotype preferentially included the HPO term “Increased mean platelet volume”; Fisher’s P = 2.10 × 10−6). All 8 probands with macrothrombocytopenia from the discovery collection had a family history suggestive of autosomal dominant inheritance. This mode of transmission was corroborated by results from cosegregation studies (Figure 1; P = 1.95 × 10−3). Systematic review of rare variants in these 8 cases within the 15 established genes implicated in macrothrombocytopenia,18 including GP1BA and GP9, did not reveal any alternative variants that could plausibly explain the platelet phenotype. We searched for further cases in the validation collection and identified 4 further probands in the ThromboGenomics data set and 6 further probands in the Japanese collection who carried a variant specific to people of Japanese ancestry (Figure 1).
Graphic representation of the 18 pedigrees with autosomal dominant inheritance of macrothrombocytopenia in the discovery and validation collections. No genome-wide excess relatedness among individuals subjected to high-throughput sequencing from different pedigrees sharing the same variant (pedigrees B and I, C and J, D and E, and H and L) could be detected by genetic analysis,25 although the shared variants in GP1BB may have been co-inherited from a distant common ancestor. Filled symbols, macrothrombocytopenia; gray symbols, unknown; blank symbols, normal platelet count and volume and absence of macrothrombocytes. Squares, males; circles, females; +/M, heterozygous; +/+: wild-type. A second variant encoding L175P was identified by Sanger sequencing in K-3 absent from K-6, which might influence PLT and MPV by affecting the cytoskeleton. The probability of the genotyping results under the null hypothesis of random segregation, conditional on the genotypes of the index cases, is 0.59 = 1.95 × 10−3.
Graphic representation of the 18 pedigrees with autosomal dominant inheritance of macrothrombocytopenia in the discovery and validation collections. No genome-wide excess relatedness among individuals subjected to high-throughput sequencing from different pedigrees sharing the same variant (pedigrees B and I, C and J, D and E, and H and L) could be detected by genetic analysis,25 although the shared variants in GP1BB may have been co-inherited from a distant common ancestor. Filled symbols, macrothrombocytopenia; gray symbols, unknown; blank symbols, normal platelet count and volume and absence of macrothrombocytes. Squares, males; circles, females; +/M, heterozygous; +/+: wild-type. A second variant encoding L175P was identified by Sanger sequencing in K-3 absent from K-6, which might influence PLT and MPV by affecting the cytoskeleton. The probability of the genotyping results under the null hypothesis of random segregation, conditional on the genotypes of the index cases, is 0.59 = 1.95 × 10−3.
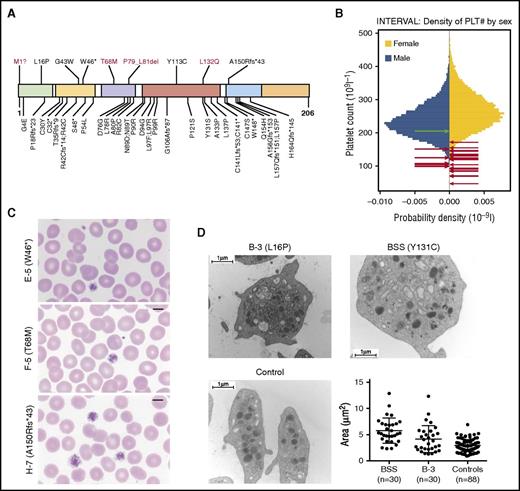
Altogether, we identified 9 unique variants in GP1BB: 1 resulting in a disruption of the canonical methionine start codon; 1 encoding a premature stop at residue 46; 5 missense variants encoding L16P, G43W, T68M, Y113C, and L132Q; a 3–amino acid deletion P79_L81del; and a frameshift in the codon for residue A150 leading to an alternative reading frame with a premature stop after 43 further residues (Figure 2A). Assessment of the potential effect of the missense and in-frame variants on GPIbβ structure, supported by prior work by McEwan et al,19 is available in supplemental Table 2 and supplemental Figure 1 on the Blood Web site. Only the variant encoding G43W is present in the ExAC database20 of more than 60 000 exome-sequenced individuals, where it is heterozygous in just 1 individual. It is noteworthy that the variant underlying the premature stop at residue 46, the variant leading to a frameshift at residue 150, and the missense variants encoding L16P, G43W, and Y113C have been previously implicated in BSS.8,10,21,22 We reason that the effects of these 5 variants in heterozygosity, except for Y113C,10,11 went unnoticed because the counts of the parents of these BSS probands were not measured or were not deemed notably low in isolation from other monoallelic cases.
(A) Localization and consequence of the 9 distinct rare variants identified in GP1BB (shown above the diagram) in the context of all 39 variants published previously, excluding the 22q11.2 deletion. Novel variants are shown in red. Known variants were obtained from a recent review,8 2 publications on BSS published since the review,26,27 and an article reporting a single case with dominant macrothrombocytopenia possibly because of a heterozygous variant encoding R42C.12 The variant encoding T68M was observed in an unaffected relative of a patient with non-BPD rare disease for whom a complete blood count was unavailable. Only the variant encoding G43W is present (in 1 individual) in the ExAC database. The domains shown in colors, from left to right, are signal peptide (1–25), LRR N-terminal (27–55), LRR (60–83), LRR C-terminal (89–143), transmembrane domain (148–172), and cytoplasmic domain (173–206). (B) Gender-stratified histograms of platelet count measurements obtained using a Sysmex hematology analyzer from 48 345 blood donors from the INTERVAL randomized controlled trial28 after adjustment for technical artifacts. The red arrows superimposed on the histograms indicate the gender and values for patients with 1 of the variants in GP1BB reported herein. For individuals I-3 and L-3, with a range of platelet counts, the midpoints were used. The green arrow indicates the gender and value for individual E-1, who is homozygous for the wild-type allele. Any superimposed arrows have been nudged slightly along the vertical axis so they can be distinguished. (C) Images of blood films from cases E-5, F-5, and H-7, with different variants in GP1BB (encoding W46*, T68M, and A150Rfs*43). Original magnification, ×100; May-Grünwald-Giemsa stain. (D) Images show representative electron micrographs of platelets from patient B-3, a patient with BSS (homozygous for a variant encoding Y131C) and an unaffected control, whereas the dot plot shows area measurements of 30 randomly selected platelets from a patient with BSS, 30 from case B-3, and 88 from 3 controls (30+30+28 platelets) with similar distributions. There was a significant difference among the means of the 3 groups (P < .0001, one-way analysis of variance). There was a significant difference between the mean area of platelets from the BSS case and patient B-3 (P = .0021), between the BSS case and controls (P < .0001), and between B-3 and controls (P = .004; Tukey’s method for multiple comparisons).
(A) Localization and consequence of the 9 distinct rare variants identified in GP1BB (shown above the diagram) in the context of all 39 variants published previously, excluding the 22q11.2 deletion. Novel variants are shown in red. Known variants were obtained from a recent review,8 2 publications on BSS published since the review,26,27 and an article reporting a single case with dominant macrothrombocytopenia possibly because of a heterozygous variant encoding R42C.12 The variant encoding T68M was observed in an unaffected relative of a patient with non-BPD rare disease for whom a complete blood count was unavailable. Only the variant encoding G43W is present (in 1 individual) in the ExAC database. The domains shown in colors, from left to right, are signal peptide (1–25), LRR N-terminal (27–55), LRR (60–83), LRR C-terminal (89–143), transmembrane domain (148–172), and cytoplasmic domain (173–206). (B) Gender-stratified histograms of platelet count measurements obtained using a Sysmex hematology analyzer from 48 345 blood donors from the INTERVAL randomized controlled trial28 after adjustment for technical artifacts. The red arrows superimposed on the histograms indicate the gender and values for patients with 1 of the variants in GP1BB reported herein. For individuals I-3 and L-3, with a range of platelet counts, the midpoints were used. The green arrow indicates the gender and value for individual E-1, who is homozygous for the wild-type allele. Any superimposed arrows have been nudged slightly along the vertical axis so they can be distinguished. (C) Images of blood films from cases E-5, F-5, and H-7, with different variants in GP1BB (encoding W46*, T68M, and A150Rfs*43). Original magnification, ×100; May-Grünwald-Giemsa stain. (D) Images show representative electron micrographs of platelets from patient B-3, a patient with BSS (homozygous for a variant encoding Y131C) and an unaffected control, whereas the dot plot shows area measurements of 30 randomly selected platelets from a patient with BSS, 30 from case B-3, and 88 from 3 controls (30+30+28 platelets) with similar distributions. There was a significant difference among the means of the 3 groups (P < .0001, one-way analysis of variance). There was a significant difference between the mean area of platelets from the BSS case and patient B-3 (P = .0021), between the BSS case and controls (P < .0001), and between B-3 and controls (P = .004; Tukey’s method for multiple comparisons).
The count and mean volume of platelets of the 18 probands were 107.9 × 109/L (range, 47–172 × 109/L) and 12.74 fL (range, 10.7–14.3 fL), respectively (Figure 2B; supplemental Table 1). In several cases, May-Grünwald-Giemsa–stained blood smears revealed platelet anisocytosis, with a subset of platelets being abnormally large; however, blood films were not available for all cases (Figure 2C; supplemental Table 1). Electron micrographs of the platelets of B-3 show significantly increased platelet surface area that is intermediate between healthy controls and BSS cases, but includes a small number of giant platelets (Figure 2D). Thus, the mean platelet volumes obtained by automated complete blood count analysis may obscure an increased spread of platelet volumes. Bleeding diathesis, including menorrhagia, epistaxis, spontaneous bleeding, and postpartum bleeding, was reported in 9 of 23 females but in none of the 8 males for whom platelet count and bleeding phenotype information was available, which suggests that the overall propensity to bleeding is, at most, only marginally increased in these patients relative to the general population. There was a reduction of at least 30% of GPIbα on platelets, as measured by flow cytometry in cases from 8 of the 9 families for which measurements were available (supplemental Table 1).
Each VWF receptor complex has 4 GPIbβ molecules, which are covalently bonded with 2 copies of GPIbα through cysteines at residues 147 and 526/527, respectively,23 and nonconvalently paired with 2 molecules each of GPV and GPIX.24 This stoichiometry may explain why a single allele encoding a mutated GPIbβ molecule may exert a dominant negative effect on the function of the VWF complex. It could also exacerbate haploinsufficiency in the cases with disruption of the methionine start codon and truncation at residue 46. It is important to note that although the overall association between monoallelic variants in GP1BB and macrothrombocytopenia is robust, it is possible that some of the 9 variants reported here are individually not causal. Therefore, further work and the continued sharing of genotype and phenotype data from many patients with macrothrombocytopenia will be required to establish the molecular consequences of these variants definitively.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank research nurse Amy Frary at the University of Cambridge and Alice Glaser at Imperial College London for their contributions to the clinical phenotyping of NIHR BioResource participants. We thank Ryoji Kobayashi, Masaaki Noguchi, Koji Miyazaki, Junichi Watanabe, Yoshiyuki Ogawa and Tomohiro Kajiguchi for providing patient samples.
M.A.L. acknowledges support from the NIHR Imperial College Biomedical Research Centre. The NIHR BioResource–Rare Diseases is funded by the National Institute for Health Research of England (award number RG65966). Research in the W.H.O. laboratory is supported by the British Heart Foundation, European Commission, Medical Research Council, NIHR, and the Wellcome Trust, and also receives support from National Health Service Blood and Transplant. C.L. and S.K.W. are supported by Medical Research Council Clinical Training Fellowships (MR/K023489/1).
Authorship
Contribution: S.S. and E.T. wrote the paper with assistance from J.C.S., M.A.L., K.F. and W.H.O.; J.C.S. maintained pedigrees, managed clinical data, and performed genotyping; S.K.W., A.M.K., C.L., P.N., K.P., D.J.P, C.R., D.P.H., R.C.T., A.D.M., NIHR BioResource, M.A.L., K.F., and S.K. provided samples and clinical data; E.G.H. conducted structure analysis; C.J.P. and K.S. managed the NIHR BioResource sequencing pipeline; S.P. coordinated the NIHR BioResource–Rare Diseases BPD project; K.D. and I.S. managed ThromboGenomics; D.P.H. supervised S.S.; S.K. managed the Japanese collection; W.J.A. analyzed the INTERVAL dataset; K.F. analyzed microscopy data; and D.G. analyzed data under the supervision of E.T.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of NIHR BioResource appears in the online appendix.
Correspondence: Ernest Turro, National Health Service Blood and Transplant, Long Rd, Cambridge CB2 0PT, United Kingdom; e-mail: et341@cam.ac.uk.
References
Author notes
S.S., S.K.W., J.C.S., and D.G. contributed equally to this study.
K.F., W.H.O., S.K., and E.T. contributed equally to this study.