Key Points
Genetic heterogeneity in non-del(5q) MDS arises within the HSPC and in committed progenitors.
Clonal selection in lineage-committed progenitors may drive the transformation to acute myeloid leukemia.
Abstract
Myelodysplastic syndromes (MDSs) are hematopoietic stem cell disorders in which recurrent mutations define clonal hematopoiesis. The origin of the phenotypic diversity of non-del(5q) MDS remains unclear. Here, we investigated the clonal architecture of the CD34+CD38− hematopoietic stem/progenitor cell (HSPC) compartment and interrogated dominant clones for MDS-initiating cells. We found that clones mainly accumulate mutations in a linear succession with retention of a dominant subclone. The clone detected in the long-term culture-initiating cell compartment that reconstitutes short-term human hematopoiesis in xenotransplantation models is usually the dominant clone, which gives rise to the myeloid and to a lesser extent to the lymphoid lineage. The pattern of mutations may differ between common myeloid progenitors (CMPs), granulomonocytic progenitors (GMPs), and megakaryocytic-erythroid progenitors (MEPs). Rare STAG2 mutations can amplify at the level of GMPs, from which it may drive the transformation to acute myeloid leukemia. We report that major truncating BCOR gene mutation affecting HSPC and CMP was beneath the threshold of detection in GMP or MEP. Consistently, BCOR knock-down (KD) in normal CD34+ progenitors modifies their granulocytic and erythroid differentiation. Clonal architecture of the HSPC compartment and mutations selected during differentiation contribute to the phenotypic heterogeneity of MDS. Defining the hierarchy of driver mutations provides insights into the process of transformation and may guide the search for novel therapeutic strategies.
Introduction
Cancer results from the sequential acquisition of genomic abnormalities that cooperate to transform normal cells. In several hematological malignancies, the initiating oncogenic event occurs in an individual stem or progenitor cell. Acute myeloid leukemias (AMLs) in particular are thought to be organized in a hierarchy with a leukemic stem cell (LSC) at its apex.1,2 Recently, 2 hierarchically organized populations with LSC activity have been identified in CD34+ AML: 1 similar to lymphoid-primed multipotent progenitors and the other having phenotypic characteristics of normal granulocyte-macrophage progenitors (GMPs).3 By contrast, hematopoietic stem cell (HSC)-derived LSCs have been demonstrated in chronic myeloid leukemia and may drive clinical relapse.4,5 Myelodysplastic syndromes (MDSs) are clinically diverse malignant disorders of aging with a propensity to evolve to AML or bone marrow (BM) failure. Evidence has been provided for the existence of an MDS-initiating cell in cases harboring a deletion of 5q.6,7 This cell resembles normal stem cells with self-renewal capacity, ability to generate differentiated cells, and capacity to engraft in immune-deficient mice. Cellular heterogeneity of cancer can be explained by the onset of mutations in a unique or several initiating cells.8 In early MDS, whole genome sequencing identifies mutations that correspond to 2 clones emerging form a unique cell, and up to 3 clones at the stage of AML.9 However, the definition and thereafter the consequences of clonal heterogeneity on the phenotypic diversity of non-del(5q) MDS is little documented.
Here we focused on studying the hierarchical organization and functionality of clones defined by molecular profiling in non-del(5q) MDS, in which we observe that the cellular and genetic complexity results from a clonal heterogeneity present in the immature hematopoietic stem/progenitor cell (HSPC) compartment and also in lineage-committed progenitors that ultimately support the transformation into AML.
Patients and methods
Human samples
BM samples were obtained from 21 patients with non-del(5q) MDS enrolled in the national trial Program Hospitalier de Recherche Clinique MDS-04 after informed consent according to ethics committee guidelines (supplemental Table 1, available on the Blood Web site). BM, cytaphereses from age-matched healthy individuals, and cord blood were used as controls.
DNA mutational analysis
A selected panel of 39 genes recurrently affected in MDS was designed (supplemental Methods) and mutations were identified by next-generation sequencing (NGS) in BM mononuclear cells (MNCs) using a PGM system (ThermoFisher Scientific, Waltham, MA) and the 384-reaction Ion AmpliSeq library kit 2.0 (Life Technologies, Chicago, IL) (supplemental Tables 2-3). DNA from colonies or single cells was obtained after lysis in water, 0.6 µM Tween 20, and 10 mg/mL proteinase K at 95°C for 1 hour and sequenced using the Sanger method.
Isolation of hematopoietic subpopulations and cell culture
CD34+ cells were enriched with a MidiMacs system (Miltenyi Biotech, Bergisch Badgach, Germany) before sorting CD34+CD38− HSPCs and CD34+CD38+ common and lineage-committed progenitors using a FACS ARIA3 (Becton Dickinson, Franklin Lakes, NJ) (see supplemental Methods and supplemental Table 4). Single HSPCs were seeded in 96-well plates on MS-5 stromal cells and cultured in H5100 MyeloCult medium (StemCell Technologies, Vancouver, Canada) with stem cell factor (SCF; 20 ng/mL), interleukin-3 (IL-3; 2 ng/mL), thrombopoietin (TPO; 10 ng/mL), and FMS-like tyrosine kinase 3 ligand (10 ng/mL) for 6 weeks. For long-term culture-initiating cell (LTC-IC) assay, CD34+ progenitors (n = 500) were cultured for 6 weeks on MS-5 in MyeloCult without cytokines. For colony-forming cell (CFC) assays, 100 CD34+CD38− cells or 1000 CD34+ cells were seeded in H4434 Methocult (StemCell Technologies) for 2 weeks. For liquid culture, CD34+ cells were expanded in Iscove modified Dulbecco medium with TPO (10 ng/mL), FLT-3L (10 ng/mL), SCF (100 ng/mL), and IL-3 (10 ng/mL) and then with erythropoietin (1 IU/mL) or granulocyte colony-stimulating factor (20 ng/mL).
Xenograft transplantation
Female 19-week-old nonobese diabetic/severe combined immunodeficiency /IL-2Rγ null (NSG) mice were irradiated with 3Gy, anesthetized before intrafemoral injection with human cells and treated with 3µg/mL buprenorphine. Mice were sacrificed after 6 to 15 weeks, and BM cells were harvested (see supplementary methods). Human cells were identified as hCD45+SSClow cells and sorted using CD34, CD14/CD15, and CD19 antibodies (supplemental Tables 5-6).
Gene expression analysis and shRNA strategy
RNA was extracted using the DNA/RNA Kit (Qiagen) and retro-transcribed by SuperScript III. Lineage-specific markers were quantified in triplicate by SYBR Green real-time polymerase chain reaction (PCR; Roche Applied Science, Penzberg, Germany). Data were analyzed using the ΔΔCt method and normalized to B2M or UBC and ACT expression (supplemental Table 7). Lentiviruses containing short hairpin RNA (shRNA) BCOR or scramble controls were purchased from Amsbio (catalog reference TL306415V) and SantaCruz (catalog references sc-72635-V and sc-108080).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5 software or open-source Statmodel package (Python software, Beaverton, OR). For 2-sample comparisons, the nonparametric Mann-Whitney test was used. Correlations were assessed by the Pearson coefficient. Depth at the variant position was used to calculate variant allele frequency (VAF), which is the proportion of mutated reads among total reads and its 95% confidence interval (CI) that give an estimated range of values.
Results
Clonal architecture in the CD34+CD38− HSPC compartment
Targeted NGS of 39 genes was used to define the mutational landscape of BM MNCs in 21 cases of non-del(5q) MDS (supplemental Tables 1-2). We found 73 mutations in 19/39 genes, with a range of 2 to 7 mutations per patient (supplemental Figure 1A-B; supplemental Table 3). Single-nucleotide polymorphism arrays did not detect CNV in these genes (not shown). We identified 53 major mutations affecting splicing factors, epigenetic regulators, chromatin modifiers, and transcriptional regulators and 20 minor mutations often affecting cohesins and signaling genes (supplemental Figure 1C-E).
CD34+CD38− HSPCs were sorted at 1 cell per well (supplemental Figure 2A) and genomic lesions were traced down to single CD34+CD38− HSPC-derived colonies obtained from cultures on MS-5 feeder in 17/21 cases. The cloning efficiency was similar between patients and controls, ruling out a specific bias of culture in patients (supplemental Figure 2B) with a mean of 31 (range, 12-78) colonies analyzed per patient. We observed an average of 2.5 molecularly distinct clones per patient (Figure 1A). Almost all mutations identified in BM MNCs were detected in the CD34+CD38− HSPCs, with the exception of STAG2L348fs in UPN6. Likewise, the proportion of mutated CD34+CD38− cells mirrored that of mutated BM MNCs (supplemental Figure 2C), in keeping with a clonal hematopoiesis derived from mutated HSPCs. Mutations accumulated in CD34+CD38− HSPCs in a linear succession in 13/17 patients. A branched clonal architecture characterized by the acquisition of 2 different mutations in 2 partially distinct subclones was observed in 4 cases (unique patient numbers [UPNs] 6, 9, 15, 17). One case (UPN 1) harbored 2 splicing mutations (SF3B1 and ZRSR2) in what appeared to be 2 independent clones, although common parental mutations in genes not assessed by our panel cannot be excluded. We identified a unique dominant clone in 16/17 patients, and UPN 2 harbored 2 codominant clones derived from a common ancestor. Nine patients exhibited a clone devoid of the identified mutations. A dominant clone containing all mutations was present in only 7/16 patients (UPNs 3, 4, 7, 8, 10, 11, 13), suggesting that early clonal dominance is inconstant in MDS.
Clonal architecture of non-del(5q) MDS CD34+CD38−HSPCs. (A) Tracking of mutations initially detected in the bone marrow of 17 MDS patients in CD34+CD38− single cell–derived colonies by the Sanger method. For each patient sample, the proportions of mutated subclone to the total number of sequenced colonies are indicated as percentages and 95% CI. Each subclone is represented as a circle. Filled circles with bold type represent the dominant or codominant subclones statistically identified using the Fisher exact test. Dominant subclones in linear architecture and in complex architecture are indicated in light and dark blue, respectively. (B) Distribution of the different mutation categories according to their respective occurrence in CD34+CD38−–derived clones. Shown are the percent of unique first events, multiple first events, and second events. (C) Mutations of different gene categories detected or not in the CD19+ cells sorted from the CD34− fraction.
Clonal architecture of non-del(5q) MDS CD34+CD38−HSPCs. (A) Tracking of mutations initially detected in the bone marrow of 17 MDS patients in CD34+CD38− single cell–derived colonies by the Sanger method. For each patient sample, the proportions of mutated subclone to the total number of sequenced colonies are indicated as percentages and 95% CI. Each subclone is represented as a circle. Filled circles with bold type represent the dominant or codominant subclones statistically identified using the Fisher exact test. Dominant subclones in linear architecture and in complex architecture are indicated in light and dark blue, respectively. (B) Distribution of the different mutation categories according to their respective occurrence in CD34+CD38−–derived clones. Shown are the percent of unique first events, multiple first events, and second events. (C) Mutations of different gene categories detected or not in the CD19+ cells sorted from the CD34− fraction.
Hierarchy of mutations in CD34+CD38− HSPCs
A unique driver mutation affecting splice (SF3B1, ZRSR2) and epigenetic regulatory (TET2, DNMT3A) genes was detected at the CD34+CD38− HSPC level in 12/17 MDS patients (Figure 1A). We found that mutations in chromatin modifiers, transcription factors, cohesin, and signal transduction genes were more frequently secondary events (Figure 1B). Part of mutations present in CD34+CD38− HSPCs were detected in CD19+ cells, including epigenetic regulatory gene (3/8) and splicing gene (3/10) mutations, underscoring their early occurrence. Other mutations were not detected, suggesting that they may preclude HSPC differentiation toward the lymphoid lineage or decrease the competitiveness of clonal B cells (Figure 1C).
Clonal evolution during the natural course of the disease
We tracked clonal evolution in the CD34+CD38− HSPCs of 2 patients over the natural course of their disease in absence of treatment. UPN 8 harbored a refractory cytopenia with multilineage dysplasia and ringed sideroblasts with a stable percentage of blasts during 30 months. Over this interval, the original SF3B1-mutated clone receded (21% to 5%), yielding dominance (52% to 95%) to an SF3B1/TET2-mutated clone (Figure 2A). UPN 3 had a refractory anemia with excess of blasts (RAEB1) followed over 113 months. At diagnosis, the initial event was a ZRSR2 mutation joined by a TET2 mutation, leading to the dominant ZRSR2/TET2-mutated clone (Figure 2B). Two years later, an additional SRSF2 mutation had been acquired in the ZRSR2/TET2-mutated cells and had driven the dominance of the triple-mutated clone. Mutations in 2 splicing genes have been detected within the same colony, suggesting a cooperative effect. Another 2 years later, the dominant clone split into 2 distinct clones defined by either a KIT or an NRAS variant. Thus, the capacity of CD34+CD38− cells to develop additional mutations over time contributes to increased clonal heterogeneity.
Evolution of clonal architecture during the natural course of the disease. Clonal architecture on the basis of single CD34+CD38− cells by the Sanger method. The proportions of each subclone are indicated as percentages and 95% CIs. Light blue circles and bold type represent the dominant subclone statistically identified using the Fisher exact test. (A) UPN 8 at diagnosis and after 30 months. (B) UPN 3 at diagnosis and after 69, 90, and 113 months.
Evolution of clonal architecture during the natural course of the disease. Clonal architecture on the basis of single CD34+CD38− cells by the Sanger method. The proportions of each subclone are indicated as percentages and 95% CIs. Light blue circles and bold type represent the dominant subclone statistically identified using the Fisher exact test. (A) UPN 8 at diagnosis and after 30 months. (B) UPN 3 at diagnosis and after 69, 90, and 113 months.
Genotyping of long-term culture-initiating cells
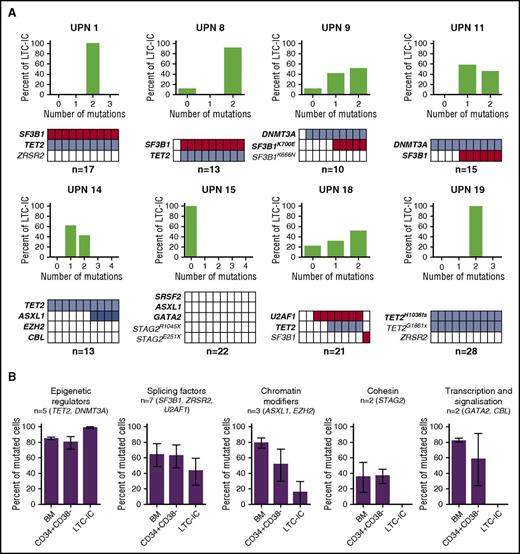
To functionally define the hematopoietic compartment targeted by mutations in MDS, we genotyped CFCs obtained from CD34+ BM cells with LTC-IC ability. The number of LTC-IC–derived CFCs was significantly decreased in MDS compared with control BM (supplemental Figure 3). A mean of 17 (range, 10-28) individual LTC-IC–derived CFCs were genotyped per patient in 8 MDS samples. Seventeen mutations were detected among the 22 mutations present in BM MNCs of the 8 patients. Zero to 3 clones were identified per sample. In 5/8 cases of early MDS (UPNs 1, 8, 9, 11, 14), the major clone was identical to the dominant clone of the CD34+CD38− compartment (Figures 1A and 3A). A major clone was also identified in 2 additional cases not previously studied in CD34+CD38− single cell–derived colonies (UPNs 18, 19). In UPN 15, a patient with RAEB2, no mutated colonies were found, whereas a dominant SRSF2/ASXL1/GATA2/STAG2E103X-mutated clone was present in most of the single CD34+CD38− HSPCs, suggesting that a majority of mutated cells could be devoid of proliferation capacity in LTC-IC assays. Other minor mutations acquired by the dominant clone in the single CD34+CD38− HSPC-derived colonies were not detected in LTC-ICs (UPNs 1, 9, 14). In 6 cases, we estimated the proportion of mutated cells in BM MNCs from VAFs9,10 and compared them with the proportions of mutated single CD34+CD38− HSPCs and of LTC-IC–derived mutated CFCs. Epigenetic and splicing factors were strongly represented in LTC-ICs (Figure 3B). This corroborates the identification of DNMT3A and TET2 or SF3B1 mutations as preleukemic or initiating events.11-15 Taken together, these in vitro data show that cells with LTC-IC activity contain initiating events corresponding to the major mutations in the CD34+CD348− compartment. However, we do not exclude that some clones were not detected as a result of low LTC-IC activity or to limited number of tested LTC-IC–derived colonies.
Frequency of mutations in LTC-IC–derived colony-forming cells. (A) Tracking of mutations in individual CFCs derived from LTC-ICs. (Upper) Proportion of LTC-ICs according to mutation burden. (Lower) Representation of the genotypes of LTC-IC–derived CFCs to 10 colonies. Each column represents 1 CFC. Colored boxes represent mutated CFCs; open boxes represent nonmutated CFCs. Each line represents 1 mutation. Number of colonies analyzed per sample is indicated. Bold type indicates major mutations. (B) Proportion of mutated cells in the BM MNC, the CD34+CD38− HSPC, and LTC-IC–derived CFCs according to variant category. Percentages of mutated cells in bulk BM MNC (BM) represent twice the VAF of heterozygous mutations on autosomes or onefold the VAF of homozygous autosomal mutations or homozygous mutations of X chromosome genes in male subjects. Percentages of mutated cells in the CD34+CD38− compartment and LTC-IC–derived CFCs represent the proportion of mutated colonies among all analyzed colonies. The numbers and type of mutations are indicated for each category. Results are expressed as means ± standard error of the mean (SEM) and compared with Student t test.
Frequency of mutations in LTC-IC–derived colony-forming cells. (A) Tracking of mutations in individual CFCs derived from LTC-ICs. (Upper) Proportion of LTC-ICs according to mutation burden. (Lower) Representation of the genotypes of LTC-IC–derived CFCs to 10 colonies. Each column represents 1 CFC. Colored boxes represent mutated CFCs; open boxes represent nonmutated CFCs. Each line represents 1 mutation. Number of colonies analyzed per sample is indicated. Bold type indicates major mutations. (B) Proportion of mutated cells in the BM MNC, the CD34+CD38− HSPC, and LTC-IC–derived CFCs according to variant category. Percentages of mutated cells in bulk BM MNC (BM) represent twice the VAF of heterozygous mutations on autosomes or onefold the VAF of homozygous autosomal mutations or homozygous mutations of X chromosome genes in male subjects. Percentages of mutated cells in the CD34+CD38− compartment and LTC-IC–derived CFCs represent the proportion of mutated colonies among all analyzed colonies. The numbers and type of mutations are indicated for each category. Results are expressed as means ± standard error of the mean (SEM) and compared with Student t test.
Clonal architecture of MDS cells with the potential to recapitulate human hematopoiesis in NSG mice
To further characterize the hierarchical origin of subclones contributing to the development of MDS, we generated patient-derived xenografts (PDXs) (supplemental Table 6). We first transplanted irradiated NSG mice with CD34+CD38− (UPN 3) or CD34+ (UPN 20) BM-derived MDS cells. Human chimerism was analyzed twice between weeks 5 and 15 after transplantation. In UPN 3, with RAEB1, human CD45+ cells represented 2.6% of the mouse BM after 6 weeks (with mainly CD14+CD15+ myeloid cells) and 0.6% after 12 weeks (with mainly CD19+ lymphoid cells). In UPN 20, with RAEB2, the engraftment with essentially lymphoid B cells was 15% and 0.3% at weeks 11 and 15, respectively (Figure 4A). Given these results, we concentrated our efforts on mice transplanted with CD3+-depleted CD45+ MDS cells at the 6- to 7-week time points, thus assessing the short-term reconstituting ability. In the 4 tested MDS cases, hCD45+ cells were detected in 10/11 mice and represented 0.3% to 20% of the mouse BM cellularity (Figure 4B). The phenotypic analysis revealed that, in 3 cases (UPNs 8, 10, 12), granulomonocytic cells were predominant, representing 39% to 66% of the hCD45+ population, whereas in 1 sample (UPN 2), CD19+ cells were dominant, as usually observed when normal cells are transplanted (supplemental Table 6).16 Sorted CD34+, CD14+CD15+, and CD19+ cells were genotyped (Figure 4C-G). In UPN 2, DNMT3AR736H and ATRXN1860S appeared as major mutations and IDH2R140Q and SF3B1K700E as minor mutations in the NSG-engrafted CD34+ cells (Figure 4C). Comparison of VAF for the 4 mutations demonstrated 2 separate clones: 1 with DNMT3A/ATRX mutations and the second with DNMT3A/ATRX/IDH2/SF3B1 mutations, consistent with their codominance in HSPCs (Figure 1A). Myeloid CD14+CD15+ cells also originated from the same 2 clones. That these CD34+ cells were able to reconstitute hematopoiesis in mice suggests that the 2 clones retained a self-renewal potential and a capacity for myeloid, but not lymphoid, differentiation. Indeed, PDX-sorted CD19+ cells did not contain the IDH2 and SF3B1 mutations, indicating that these cells originated from the DNMT3A/ATRX-mutated cells of the CD34+CD38− compartment. Interestingly, cells with the lowest mutation number had the greatest multilineage potential, being the most competitive in the in vivo xenograft context (Figure 4D). In UPN 8, the SF3B1 and TET2 mutations were detected in myeloid and lymphoid progenies (Figure 4E) as well as the SRSF2, TET2, and STAG2 mutations in UPN 12, suggesting that the dominant clone of the CD34+CD38− HSPCs with multilineage differentiation potential also reconstituted short-term hematopoiesis in murine BM. In UPN 10, the SRSF2 and TET2 mutations of the dominant clone were detected only in the CD14+CD15+ myeloid cells, although CD19+ could not be detected because they did not develop in the mice (Figure 4F). In UPN 12, the minor NRAS mutation acquired and identified from CD34+CD38− single HSPC-derived colonies was undetectable (Figure 4G). These results thus identified MDS cells with in vivo hematopoiesis initiating activity and show that the dominant clone found in the CD34+CD38− HSPCs is the clone that reconstitutes short-term human hematopoiesis in mice.
Xenotransplantation of MDS cells in NSG mice. (A-B) Immunophenotypic profiling of human cells in NSG mice. Shown are percentages of hCD45+, myeloid (CD14+ or CD15+), and B lymphoid (CD19+) cells in the hCD45+ cell fraction, at 5 to 15 weeks posttransplantation. (C-G) Genotype of sorted MDS CD14+CD15+, CD19+, or CD34+ populations after xenografting. Shown are VAFs and the 95% CIs of each mutation in BM MNCs, PDX CD14+CD15+, PDX CD19+, and PDX CD34+. In UPNs 2, 10, and 12, mutations were screened by NGS. For UPN 8, mutations in PDX CD14+CD15+ and CD19+ populations were detected by Sanger sequencing on amplified whole genome DNA. (D) For UPN 2, hypotheses regarding the contribution of CD34+-mutated clones to their myeloid CD14+CD15+ (My) and lymphoid CD19+ (Ly) progenies are schematically depicted. Each mutation is represented by its initial. A color spectrum is used for the different intervals of variant allele frequencies.
Xenotransplantation of MDS cells in NSG mice. (A-B) Immunophenotypic profiling of human cells in NSG mice. Shown are percentages of hCD45+, myeloid (CD14+ or CD15+), and B lymphoid (CD19+) cells in the hCD45+ cell fraction, at 5 to 15 weeks posttransplantation. (C-G) Genotype of sorted MDS CD14+CD15+, CD19+, or CD34+ populations after xenografting. Shown are VAFs and the 95% CIs of each mutation in BM MNCs, PDX CD14+CD15+, PDX CD19+, and PDX CD34+. In UPNs 2, 10, and 12, mutations were screened by NGS. For UPN 8, mutations in PDX CD14+CD15+ and CD19+ populations were detected by Sanger sequencing on amplified whole genome DNA. (D) For UPN 2, hypotheses regarding the contribution of CD34+-mutated clones to their myeloid CD14+CD15+ (My) and lymphoid CD19+ (Ly) progenies are schematically depicted. Each mutation is represented by its initial. A color spectrum is used for the different intervals of variant allele frequencies.
Clonal architecture at the level of clonogenic myeloid progenitors
We then hypothesized that genetic landscape can evolve along hematopoietic differentiation. To address this question specifically at the progenitor level, CD34+CD38− HSPCs were seeded in methylcellulose for 14 days with a mean cloning efficiency of 24% (range, 17-45). A mean of 22 (range, 12-38) CFCs was genotyped in 7 MDS patients. All clones identified in single CD34+CD38−–derived colonies were also present in CFCs in 4 cases (UPNs 3, 6, 8, 16), whereas CFCs with an isolated ZRSR2 mutation in UPN 1, or CFCs devoid of recurrent mutations in UPNs 14 and 15 were undetectable (Figure 5A). The proportions of each clone did not vary significantly in 3 cases (UPNs 3, 8, 16). In contrast, in 4 cases (UPNs 1, 6, 14, 15), the clonal distribution was different between CD34+CD38− cells and CFCs. In UPN 6, the minor STAG2R1045X-mutated clone amplified at the expense of the dominant BCOR-mutated clone. In UPN 14, a minor clone containing EZH2 and CBL variants became the dominant clone in CFCs. Last, in UPN 15, a STAG2R1045X-mutated clone decreased to the benefit of the STAG2E251X-mutated clone; thus, although the repartition of clones appeared to be very similar between single-cell colonies and CFCs issued from CD34+CD38− HSPCs in some MDS cases, arguing that myeloid expansion starts in the HSPC compartment, changes in clonal architecture between CD34+CD38−–derived single-cell colonies and clonogenic progenitors reveal that clones with specific mutations may experience lineage- and differentiation state–dependent selective pressure.
Genotyping of MDS progenitors. (A) Genotyping of CD34+CD38− HSPC-derived CFCs. One hundred sorted bone marrow CD34+CD38− cells were seeded in methylcellulose for CFCs of 7 MDS patients. Mutations identified in bulk BM MNCs were tracked in individual colonies. Clones are defined as colonies with the identical patterns of mutations. Proportions of clones derived from individual CD34+CD38− cells and in CFCs derived from CD34+CD38− cells are compared and represented by bars showing 95% CIs. (B) Immunophenotypic quantification of CMP, GMP, and MEP in the CD34+CD38+ cell compartment for 8 MDS patients and 1 cytapheresis used as control (see supplemental Methods). Density plots are shown and percentages of each studied cell population are indicated. (C) Genotyping of sorted CMPs, GMPs, and MEPs from 8 MDS patients by Sanger sequencing (200 cells per sample) or by NGS (UPN 21). Colored boxes correspond to detected mutation and open boxes for undetected mutation.
Genotyping of MDS progenitors. (A) Genotyping of CD34+CD38− HSPC-derived CFCs. One hundred sorted bone marrow CD34+CD38− cells were seeded in methylcellulose for CFCs of 7 MDS patients. Mutations identified in bulk BM MNCs were tracked in individual colonies. Clones are defined as colonies with the identical patterns of mutations. Proportions of clones derived from individual CD34+CD38− cells and in CFCs derived from CD34+CD38− cells are compared and represented by bars showing 95% CIs. (B) Immunophenotypic quantification of CMP, GMP, and MEP in the CD34+CD38+ cell compartment for 8 MDS patients and 1 cytapheresis used as control (see supplemental Methods). Density plots are shown and percentages of each studied cell population are indicated. (C) Genotyping of sorted CMPs, GMPs, and MEPs from 8 MDS patients by Sanger sequencing (200 cells per sample) or by NGS (UPN 21). Colored boxes correspond to detected mutation and open boxes for undetected mutation.
Hierarchical organization of mutations in common and committed progenitors
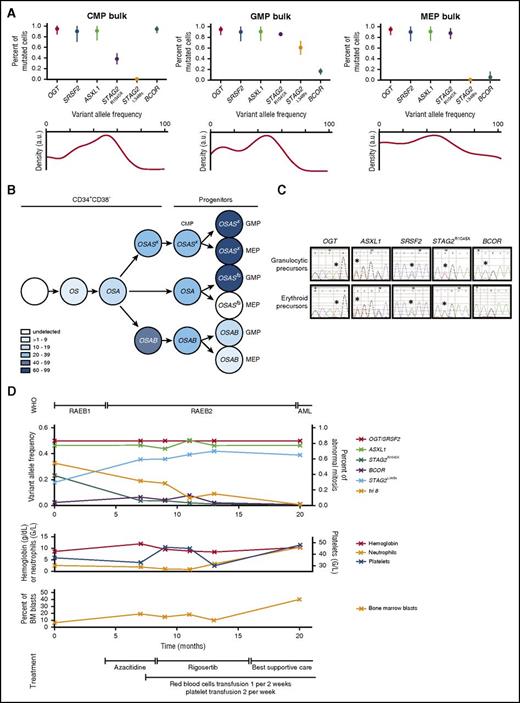
We further investigated the hierarchy of mutations in sorted common myeloid progenitors (CMPs), GMPs, and megakaryocyte-erythroid progenitors (MEPs) from the CD34+CD38+ populations of 8 MDS BM samples (see supplemental Methods). Intrinsic potentials of these progenitors were confirmed in CFC assays and by expression of lineage-restricted markers (supplemental Figure 4A-B). As expected,7,17 the percentage of MEPs among CD34+CD38+ cells was low and the percentage of CMPs and GMPs was high in RAEB1/2 cases (UPNs 3, 6, 14, 21) (Figure 5B). In contrast, in 3 MDSs with ringed sideroblasts (UPNs 1, 4, 8), the percentage of MEPs was preserved, consistent with BM enrichment of erythroblasts.
On a genetic level, 32/33 mutations present in bulk marrow were detected in sorted subpopulations (Figure 5C). All mutations of BM MNCs were rediscovered in each population of UPN 3, 4, 8, 14, and 21. In UPN 1, a ZRSR2 mutation detected in CD34+CD38− HSPCs (Figure 1A), and none of CFCs (Figure 5A), was undetectable in CMPs, GMPs, and MEPs. In UPN 15, the dominance of an ASXL1/SRSF2/GATA2/STAG2E251X clone over an ASXL1/SRSF2/GATA2/ STAG2R1045X clone among CFCs was manifested by amplification of a STAG2E251X mutation in GMPs and MEPs, in concert with diminution of a STAG2R1045X mutation to beneath the detection threshold in GMPs. In UPN 6,with 1 mutation affecting the BCOR gene, observed at high frequency in CD34+CD38− cells, was detectable in CMPs and but not in GMPs or MEPs; this is also coincident with low detection in CFCs. This BCOR mutation appeared irreconcilable with a commitment to differentiation (Figure 5A). Furthermore, a STAG2L348fs mutation, not detected in either the CD34+CD38− compartment or CFCs (Figures 1A and 5A), was undetectable in the CMP and MEP, and appeared for the first time in the GMP population (Figure 5C). We confirmed that BCOR, STAG2R1045X, and STAG2L348fs mutations did not coexist in the same cell by NGS (Figure 6A) and by single CMP, GMP, and MEP genotyping (supplemental Figure 5). An OGT/SRSF2/ASXL1/BCOR-mutated clone was dominant in CMPs. The MEPs contained 2 clones, including 1 dominant OGT/SRSF2/ASXL1/STAG2R1045X-mutated clone; the GMPs contained 2 codominant clones harboring STAG2R1045X or STAG2L348fs mutations (Figure 6B). BCOR became a minor mutation in GMPs and MEPs (Figure 6A; supplemental Figure 5). A STAG2L348fs mutation was detectable only in bulk or in individual GMPs, although we could not exclude the presence of STAG2L348fs in self-renewing compartments such as HSC or multipotent progenitor (MPP). To test this hypothesis, we genotyped sorted the HSCs, MPPs, CMPs, GMPs, and MEPs of UPN 21. Lineage marker depletion using CD3, CD14, and CD19 did not modify the proportion of each subpopulation (supplemental Figure 6A-B). Among 7 mutations—including 4 major STAG2F798fs, ETV6, U2AF1, and ASXL1 and 3 minor NRAS, STAG2A916fs, and STAG2R1045X mutations identified by NGS—the 2 minor STAG2A916fs and STAG2R1045X mutations were not detectable in the bulk HSC but were detected with a VAF of 6% and 11% in CMP, respectively (supplemental Figure 2A,C). By direct Sanger sequencing of single HSC or CMP, we detected 5/102 (5%) STAG2R1045X-mutated HSCs and 8/73 (11%) STAG2R1045X-mutated CMPs (supplemental Figure 6B,D). We conclude that a rare mutation detected by single-cell sequencing in the HSC compartment may amplify downstream in the context of myeloid differentiation.
Clonal evolution with hematopoietic differentiation. (A) Targeted resequencing of sorted CMPs, GMPs, and MEPs by NGS for UPN 6 (200 cells per fraction). (Upper) Proportion of mutated cells represents twice the VAF, determined by NGS, of heterozygous mutations in autosomes and onefold the VAF of mutations of a gene located on the X chromosome in this male subject. Bars represent 95% CIs. (Lower) Kernel density plot of VAFs determined by NGS representation facilitating inference of the presence and genetic composition of subclones (SiClone). (B) Schematic representation of the clonal architecture of CD34+CD38− hematopoietic stem cell and progenitor compartments at diagnosis. Each mutation is represented by its initial. A color spectrum is used for the different intervals of variant allele frequencies. (C) Mutation pattern of granulocytic and erythroid precursors. Patient (UPN 6)-sorted CD34+ cells were expanded in vitro and transitioned to granulocytic or erythroid differentiation. Mutations were tracked by Sanger sequencing in granulocytic and erythroid differentiated cells (*point mutations). (D) Clonal evolution of UPN 6 over 20 months. World Health Organization (WHO) classification, blood and bone marrow parameters, treatments, mutations, and cytogenetic abnormality are indicated.
Clonal evolution with hematopoietic differentiation. (A) Targeted resequencing of sorted CMPs, GMPs, and MEPs by NGS for UPN 6 (200 cells per fraction). (Upper) Proportion of mutated cells represents twice the VAF, determined by NGS, of heterozygous mutations in autosomes and onefold the VAF of mutations of a gene located on the X chromosome in this male subject. Bars represent 95% CIs. (Lower) Kernel density plot of VAFs determined by NGS representation facilitating inference of the presence and genetic composition of subclones (SiClone). (B) Schematic representation of the clonal architecture of CD34+CD38− hematopoietic stem cell and progenitor compartments at diagnosis. Each mutation is represented by its initial. A color spectrum is used for the different intervals of variant allele frequencies. (C) Mutation pattern of granulocytic and erythroid precursors. Patient (UPN 6)-sorted CD34+ cells were expanded in vitro and transitioned to granulocytic or erythroid differentiation. Mutations were tracked by Sanger sequencing in granulocytic and erythroid differentiated cells (*point mutations). (D) Clonal evolution of UPN 6 over 20 months. World Health Organization (WHO) classification, blood and bone marrow parameters, treatments, mutations, and cytogenetic abnormality are indicated.
Clonal selection at the level of committed progenitors can drive leukemic transformation
In UPN 6, the presence of the STAG2L348fs mutation in a branch of the GMP compartment could account for the amplification of GMPs representing 51% of CD34+CD38+ cells over MEPs representing 8% of this population (Figure 5B). Therefore, we hypothesized that the STAG2L348fs mutation may drive the evolution of the disease. To test this hypothesis, we performed NGS on follow-up samples over 20 months. We observed that the initially dominant OGT/SRSF2/ASXL1/STAG2R1045X clone diminished in parallel with a partial trisomy 8 and was overtaken by the OGT/SRSF2/ASXL1/STAG2L348fs clone that gained dominance as the disease progressed (Figure 6D). BCOR-mutated cells always remained restricted to a minor clone. This argues compellingly that the blast population emerged from a GMP-derived clone in which the STAG2L348fs mutation developed and that may have acquired self-renewal capacities to drive leukemic transformation.
Inhibition of granulocytic and erythroid differentiation by BCOR invalidation
UPN 6 harbored a major truncating BCOR variant in the CD34+CD38− HSPC and CMP compartments. Its low frequency in GMPs, MEPs, and CFCs suggests that BCOR expression may be critical for myeloid differentiation (Figures 5A and 6A; supplemental Figure 6A). Using a liquid culture system, UPN 6 CD34+-derived granulocytic and erythroid precursors were amplified to investigate their mutation status. OGT, SRSF2, ASXL1, and STAG2R1045X were detected in the 2 populations. In contrast, BCOR-mutated granulocytic or erythroid precursors were poorly detectable (Figure 6C). This suggests that BCOR mutations either decreased the fitness of precursors or prevented their differentiation.
Starting from normal CD34+ cells, we then observed that BCOR messenger RNA expression increased during granulocytic and erythroid differentiation (Figure 7A). Upon KD of endogenous BCOR protein by shRNAs (Figure 7B), BCOR low-expressing cells displayed increased proliferative capacity with SCF, IL-3, IL-6, and TPO for 18 days (Figure 7C; supplemental Figure 7), whereas the number of granulocytic and erythroid CFCs generated was significantly diminished, suggesting a differentiation arrest (Figure 7D). BCOR KD erythroid cells had low GPA expression and retained high CD49d expression at day 18 (Figure 7E, left). Moreover, they expressed a lower level of GATA1 and HBB transcripts, suggesting that erythroid differentiation was delayed (Figure 7E, right). BCOR KD granulocytic cells expressed high levels of MPO and variable levels of SPI1 depending on the shRNA used, and they lost CD33 expression more rapidly than control scrambled shRNA (shSCR) cells suggesting, that decreased BCOR expression may accelerate granulocytic differentiation (Figure 7F). Together, these results show that functional inactivation of BCOR modifies granulocytic and erythroid differentiation and promotes CD34+ cell expansion.
Impact of BCOR on erythroid and granulocytic differentiation. (A) Evolution of BCOR gene expression during normal granulocytic and erythroid differentiation. BCOR messenger RNA level as measured by reverse transcriptase quantitative PCR (RT-qPCR) is shown as normalized relative quantities (NRQs) ± SEM to B2M and ACT. (B) shRNA-mediated BCOR silencing in normal human CD34+ progenitors (day 7). shSCR as control. Western blot to BCOR; actin is used as loading control. (C) Proliferation of short hairpin BCOR (shBCOR) or shSCR human CD34+ progenitors was driven by IL-3, SCF, FLT-3L, and TPO for 18 days. Results are representative of 4 independent experiments. (D) Clonogenic progenitor growth. shBCOR or shSCR CD34+ progenitors were amplified in liquid culture for 6 days and seeded in methylcellulose. Colony numbers ± SEM. (E) (Left) Immunophenotypic quantification of CD71, GPA, Band3, and CD49d expression in shBCOR and shSCR cells at day 18 of erythroid differentiation. Biparametric histograms are shown and percentages are indicated. Results representative of 3 independent experiments. (Right) GATA1 and HBB gene expression according to expression of shBCOR or shSCR by RT-qPCR. Results expressed as NRQs to UBC and ACT ± SEM represent 2 independent experiments in duplicates. (F) (Left) Immunophenotypic quantification of CD33 and CD11b. (Right) SPI1 and MPO gene expression by RT-qPCR.
Impact of BCOR on erythroid and granulocytic differentiation. (A) Evolution of BCOR gene expression during normal granulocytic and erythroid differentiation. BCOR messenger RNA level as measured by reverse transcriptase quantitative PCR (RT-qPCR) is shown as normalized relative quantities (NRQs) ± SEM to B2M and ACT. (B) shRNA-mediated BCOR silencing in normal human CD34+ progenitors (day 7). shSCR as control. Western blot to BCOR; actin is used as loading control. (C) Proliferation of short hairpin BCOR (shBCOR) or shSCR human CD34+ progenitors was driven by IL-3, SCF, FLT-3L, and TPO for 18 days. Results are representative of 4 independent experiments. (D) Clonogenic progenitor growth. shBCOR or shSCR CD34+ progenitors were amplified in liquid culture for 6 days and seeded in methylcellulose. Colony numbers ± SEM. (E) (Left) Immunophenotypic quantification of CD71, GPA, Band3, and CD49d expression in shBCOR and shSCR cells at day 18 of erythroid differentiation. Biparametric histograms are shown and percentages are indicated. Results representative of 3 independent experiments. (Right) GATA1 and HBB gene expression according to expression of shBCOR or shSCR by RT-qPCR. Results expressed as NRQs to UBC and ACT ± SEM represent 2 independent experiments in duplicates. (F) (Left) Immunophenotypic quantification of CD33 and CD11b. (Right) SPI1 and MPO gene expression by RT-qPCR.
Discussion
In the present study of non-del(5q) MDS, we characterized the first genetic hits that initiate disease, identifying a dominant clone among the CD34+CD38− HSPCs. This molecularly defined clone exhibits LTC-IC activity and reconstitutes human short-term hematopoiesis in NSG mice. Rare mutations in HSCs that are distinct from LTC-ICs may also amplify in HSPCs. Contributing to tumor heterogeneity, mutations that expand in lineage-committed progenitors may drive AML transformation.
Leukemia-initiating cell identification depends upon self-renewal capacity either intrinsic to a stem cell or acquired in a progenitor as a result of genetic insults.2,3 Here, we identify the MDS-initiating events within LTC-ICs in early, but not in advanced, MDS. In the latter, mutations may either decrease the fitness of LTC-ICs or arise downstream of this compartment. This observation is consistent with the identification of lymphoid-primed multipotent progenitors and GMP-like progenitors, such as LSCs in AML.3 LTC-ICs are less frequent in dysplastic compared with normal marrow,6 which may reflect dependency on the stromal niche.18 Consistently, coinjection of autologous mesenchymal stromal cells augments engraftment efficiency of MDS cells difficult to expand in immune-deficient mice.7,18-21 Here, we successfully used a strategy of T-lymphocyte depletion of CD45+ MNCs. Results reveal a myeloid differentiation bias that could be related to impaired lymphopoiesis consistent with the involution of B-cell progenitors in MDS BM22 or to positive selection of a myeloid-biased multipotent progenitor. We cannot exclude that progenitors mainly of myeloid origin rather than HSCs may have engrafted because we analyzed engraftment efficiency 6 to 7 weeks posttransplant. That said, the engraftment of CD34+ cells belonging to the dominant clone demonstrates their self-renewal capacity. MDS-initiating cell that engrafts in mice belongs to the dominant clone of the CD34+CD38− compartment, which contains the founding mutations. Long-term engraftment of del(5q) or SF3B1-mutated cells derived from HSCs has been also reported.7,15 In contrast, xenografts in AML can be restricted to a subclone distinct from the one that emerges at relapse, whereas T-cell acute lymphoblastic leukemia xenografts usually bear genetic resemblance to relapse samples.23-25
B or T lymphocytes have already been reported to participate in clonal MDS hematopoiesis.18,26 Here, B lymphocytes display a mutational repertoire limited to epigenetic or splicing gene mutations. This observation implicates several hypotheses: (1) mutations in other genes may preclude the transition of HSCs to lymphoid progenitors or impair the fitness of mutant B cells; (2) secondary mutations may appear in a CD34+CD38− multipotent progenitor subset with low lymphoid potential27,28 ; or (3) mature B-cell progeny may originate from long-lived lymphoid progenitor with low proliferative capacity coming from a CD34+CD38− clone harboring only early genetic lesions.29-31 Furthermore, we show that myeloid compartments exhibit distinct combinations of mutations that reflect spatial heterogeneity of BM, as in solid tumors8 ; for instance, STAG2L348fs amplifies in GMPs whereas BCOR mutations become poorly detectable in GMPs and MEPs.
The BCOR gene is mutated in MDS, AML, and aplastic anemia, and truncating mutations of BCOR result in the downregulation of BCOR transcript.32-34 Bcor−/− mouse embryonic stem cells leads to reduced lymphocytes and erythrocytes, and in a murine model of Bcor loss-of-function mutation, adult progenitor cell proliferation and its granulomonocytic differentiation are enhanced.35,36 Here, we used shRNAs to inactivate BCOR, which limits progenitor differentiation and modulates granulocytic and erythroid maturation in vitro. Thus, we suggest a role for BCOR mutations in the development of cytopenias.
In conclusion, we propose a model in which the clonal architecture of the HSPC compartment and various mutations interacting throughout differentiation may contribute to the phenotypic heterogeneity of MDS. Defining the architecture and hierarchy of driver mutations sheds light on the process of leukemic transformation. Therapeutic decisions must reconcile the dual goals of targeting the dominant clone in the HSPC compartment as well as eradicating subclones with mutations that are likely to drive transformation.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Audrey Gauthier and Marlène Dejean for technical assistance and Rosa Sapena for recording clinical data; M. Vidaud and staff of the next-generation sequencing platform Assistance Publique-Hôpitaux de Paris/INSERM of Cochin Hospital; C. Preudhomme from the Laboratory of Hematology, Centre Hospitalier Regional Universitaire of Lille; V. Bardwell at University of Minnesota (Minneapolis, MN) for antibody to BCOR; and MA Murakami, from the Dana-Farber Cancer Institute, Boston, MA, for critical review of the manuscript. NSG mice were carefully handled by the Commissariat à l’Energie Atomique and Charles River teams in the Institut de recherche en Radiobiologie Cellulaire et Moléculaire; we thank J. Tilliet and I. Rombeau headed by C. Joubert and Y. Moreau.
This work was supported by grants from INSERM, the Institut National du Cancer and the Direction Générale de l’Offre de Soins of the French Ministry of Social Affairs and Health through the Programme Hospitalier de Recherche Clinique (grant PHRC MDS-04), the Ligue Nationale Contre le Cancer (Equipes labellisées) (F.P. and O.A.B.), the Commissariat à l’Energie Atomique, the Universities Paris Descartes, Paris 7/Diderot and Paris Saclay, and the association Laurette Fugain (O.A.B.), and by fellowship grants from the Ministère de l’Enseignement supérieur et de la Recherche (France) and from the Société Française d’Hématologie (V.C.) and the Site de Recherche Intégrée sur le Cancer, Cancer Research for Personalized Medicine (O.K.).
Authorship
Contribution: M.F., F.P., V.C., and O.K. designed the study; V.C., M.-L.A., C.D., A.R., H.G., S.B., M.C., and C.L. performed the experiments; L.W., D.B., N.C., L.L., and S.R. recorded patients and provided samples and clinical data; V.C., M.-L.A., O.K., M.D., E.L., and S.R. analyzed and interpreted the data; E.L. and O.A.B. critically reviewed the manuscript; and O.K., M.F., and F.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michaela Fontenay, Service d’hématologie biologique, Hôpital Cochin, 27 rue du Faubourg Saint-Jacques, 75014 Paris, France; e-mail: michaela.fontenay@inserm.fr; Françoise Pflumio, INSERM Unité Mixte de recherche 967, Commisariat à l'Energie Atomique, 10 route du Panorama, BP6, 92265 Fontenay-aux-Roses Cedex, e-mail: francoise.pflumio@cea.fr; and Olivier Kosmider, Service d’hématologie biologique, Hôpital Cochin, 27 rue du Faubourg Saint-Jacques, 75014 Paris, France; e-mail: olivier.kosmider@aphp.fr.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal