Key Points
GLI3R inhibits Hh signaling and is required for response to SMO antagonist in AML.
GLI3 is silenced in AML, and decitabine restores GLI3 expression and leads to modulation of Hh signaling.
Abstract
The Hedgehog (Hh) signaling pathway is activated in many cancers and is a promising target for therapeutic development. Deletions in the receptor Patched (PTCH) or activating mutations in Smoothened (SMO) have been reported in basal cell carcinoma and medulloblastoma, but are largely absent in most tumor types. Therefore, the mechanism of pathway activation in most cancers, including hematological malignancies, remains unknown. In normal tissues, Hh pathway activation via PTCH/SMO causes an increase in the downstream transcriptional activator GLI1 and a decrease in the GLI3 transcriptional repressor (GLI3R). In this article, we confirm that the Hh pathway is active in acute myeloid leukemia (AML), however, this activity is largely independent of SMO. Epigenetic and gene expression analysis of The Cancer Genome Atlas AML data set reveals that GLI3 expression is silenced in most AML patient samples, and the GLI3 locus is abnormally methylated. We show that GLI3R is required for the therapeutic effect of SMO antagonists in AML samples and restoration of GLI3R suppresses the growth of AML. We additionally demonstrate that GLI3R represses AML growth by downregulating AKT expression. In summary, this study provides the first evidence that GLI3R plays an essential role in SMO-independent Hh signaling in AML, and suggests that GLI3R could serve as a potential biomarker for patient selection in SMO antagonist clinical trials. Furthermore, these data support rational combinations of hypomethylating agents with SMO antagonists in clinical trials.
Introduction
The Hedgehog (Hh) signaling pathway plays a critical role in embryonic development and adult tissue homeostasis.1 The Hh pathway involves 3 Hh ligands, Sonic (SHh), Indian, and Desert, that activate the signaling cascade by binding to the membrane receptor Patched (PTCH).2 In the absence of the ligand, PTCH represses pathway by inhibiting the activity of Smoothened (SMO).3 In the presence of Hh ligand, the inhibitory effects of PTCH on SMO are relieved, and SMO initiates a signaling cascade that is mediated by members of the GLI family (GLI1, GLI2, and GLI3) transcription factors.4 GLI1 functions as a critical transcriptional activator of the Hh pathway, and its function is reinforced by a positive feedback loop, because GLI1 is also a target gene of Hh signaling.5,6 GLI2 and GLI3 exist in both full-length forms as transcriptional activators and in proteolytically processed forms as transcriptional repressors. Processing of GLI2/GLI3 is regulated by Hh signaling activity, and in the absence of Hh ligand, both GLI2 and GLI3 exist as transcriptional repressors (GLI2R and GLI3R). Although processing of GLI2 is highly variable and tissue context dependent, GLI3 processing is complete in most tissues and therefore serves exclusively as a strong repressor of Hh pathway target genes. Loss of GLI3R-mediated suppression of Hh targets is sufficient to activate the pathway independent of Hh ligands or SMO.7-9 Because both GLI activator and GLI repressor forms share the same DNA binding motifs, the proteins compete with each other for access to the GLI binding sites in target genes; thus, the overall output of the Hh pathway is dictated by a balance between the transcriptional activity of the activator and repressor forms of GLI proteins.10 In addition to target genes like PTCH1, GLI1, and HHIP, which are members of the Hh signaling network, the Hh pathway also controls other oncogenic pathways by regulating target genes, such as BCL-2, AKT, and CCND1.11
Hh signaling is aberrantly activated in many tumor types, including leukemia, making it an attractive target for cancer therapy.12 Approaches to inhibit Hh signaling for therapeutic benefit have focused primarily on SMO inhibitors (SMOis), and the identification of cyclopamine, a plant-derived SMOi, provided initial evidence that pathway activity could be pharmacologically modulated.13-15 Since then, several SMO antagonists have been identified through small-molecule screens, and 2 such agents, vismodegib and sonidegib, are currently approved by the FDA for the treatment of basal cell carcinoma (BCC). These and other SMOis, including Pfizer’s glasdegib (PF-04449913),16 are currently being studied in various hematologic malignancies.
The rationale for using SMOi in leukemia comes from 3 types of evidence. The first type of evidence comes from gene profiling experiments that have demonstrated that the expression of GLI genes is associated with poor clinical outcomes.17 The second type evidence comes from genetically engineered mouse models of leukemia, such as BCR-ABL–driven acute leukemia or a FLT3-ITD model of myeloproliferative disease, where SMO was shown to be critical for disease progression.18,19 The third type of evidence supporting the use of SMOis in leukemia comes from clinical trials. In a phase 1 study of PF-04449913, encouraging single-agent activity was noted, with 49% of patients experiencing reductions in marrow blasts and 1 patient achieving a complete response.16 A phase 1 study combining the hypomethylating agent 5-azacitadine and sonidigeb showed promising activity, including patients refractory to 5-azacitadine alone.20 However, attempts to demonstrate Hh target gene modulation in primary acute myeloid leukemia (AML) blasts treated with SMOi have been largely unsuccessful, and others have reported no role for SMO in leukemia models, leaving the matter far from settled.21-23 Responses to SMOis are not universal, and currently there are no biomarkers to select patients who are more likely to respond to SMOis.
In this study, we show that Hh signaling is active in AML, but is largely independent of SMO and therefore inherently resistant to SMOis. We also demonstrate that GLI3 behaves as a tumor suppressor in AML and is a key regulator of Hh target gene expression, specifically AKT. Finally, we show that GLI3R expression can be modulated by hypomethylating agents and should be further investigated as a biomarker for patient selection and/or response in SMOi clinical trials.
Materials and methods
Cells and reagents
Human myeloid leukemic cell lines were cultured in RPMI 1640 medium, supplemented with 10% fetal bovine serum and 2 mM l-glutamine. Clinical specimens were obtained from patients with newly diagnosed AML and healthy donors. All human subjects provided informed consent, and the study was approved by the University of Southern California Institutional Review Board. Mononuclear cells were isolated by density centrifugation by using Ficoll-Paque Plus (GE Healthcare Bio-Sciences, Pittsburgh, PA). Decitabine was procured from Sigma. The SMO antagonist glasdegib (PF-04449913) was obtained from Pfizer.
Treatments and transfections
Cells were cultured in advanced RPMI 1640 with 1% fetal bovine serum in the presence and absence of PF-04449913 for 48 hours before harvesting for additional analysis.
The Myc-GFP–tagged GLI3R construct was a gift from James Briscoe (MRC National Institute for Medical Research, London, United Kingdom). K562 and KG1a cells were transfected with Gli3R vector, and medium containing 100 nM PF-04449913 was replaced at 24 hours and harvested at 48 hours. Rescue experiments were done by using a constitutively active pcDNA3 Myr-HA-AKT1 construct from Addgene (Cambridge, MA).
For knockdown studies, cells were transiently transfected with 50 nM GLI1 and SMO or nonspecific (NS) small interfering (siRNA) (Santa Cruz Biotechnology, Santa Cruz, CA) by using Lipofectamine RNAimax in accordance with the manufacturer's instructions. Analysis was done after 48 hours. Silencing of GLI3 was performed by using a third-generation lentiviral-based siRNA transduction system. The targeted sequence of the 4 iLenti-si-Gli3 is provided in supplemental Methods, available on the Blood Web site.
Cell viability assay
Cell viability was measured by using a trypan blue dye exclusion assay. Cells were incubated with equal amounts of trypan blue for 5 minutes and counted on a hemocytometer by using a microscope.
Real-time quantitative polymerase chain reaction
The expression of Hh pathway components was measured by real-time quantitative polymerase chain reaction (Q-PCR) by using Fast Taqman reagents (Applied Biosystems) as described in supplemental Methods.
Western blotting
Cells were lysed in cold radioimmunoprecipitation assay lysis buffer containing protease inhibitors (Complete; Roche Applied Science). Full details of western blotting are provided in supplemental Methods.
GLI reporter assay
GLI reporter activity in leukemia cell lines was measured by using GLI-responsive firefly luciferase vector, and the details are provided in supplemental Methods.
Chromatin immunoprecipitation Q-PCR
A chromatin immunoprecipitation (ChIP) assay was done by using the LowCell ChIP kit (Diagenode, Denville, NJ), and the details of ChIP are provided in supplemental Methods.
Analysis of gene expression, mutations, and methylation in the TCGA AML data set
TCGA AML cohort DNA methylation, microarray data,24 and normal progenitors from Jung et al25 were processed and normalized by using the minfi package.26 For RNA sequencing–based transcript-level expression quantification, we used Kallisto27 to perform pseudoalignment and Arkas (http://dx.doi.org/10.1101/031435) to normalize, cluster, and plot the data. Plots were generated by using R (https://www.r-project.org) and Bioconductor.28
Murine xenograft models
K562 cells were transfected with a virus containing the firefly luciferase and green fluorescent protein genes (pMSCV-luc-IRES-GFP). One million GFP+ cells were injected IV into NOD-SCID-γc−/− mice (The Jackson Laboratory) bred in-house under an approved institutional animal care and use committee protocol. Mice were treated with vehicle or PF-04449913 (20 mg/kg) by oral gavage and decitabine (DAC, 0.25 mg/kg) by intraperitoneal injection once daily for 5 days with 2 days off for up to 28 days for all treatment groups. Engraftment was confirmed by using bioluminescent imaging.
Statistical analysis
Comparisons between treatments were performed by using a two-tailed, paired Student t test. For all analyses, P < .05 was considered statistically significant. Survival curves (Kaplan-Meier) were generated by using GraphPad Prism software, and significance was tested by using the log-rank test.
Results
Hh signaling is active in AML
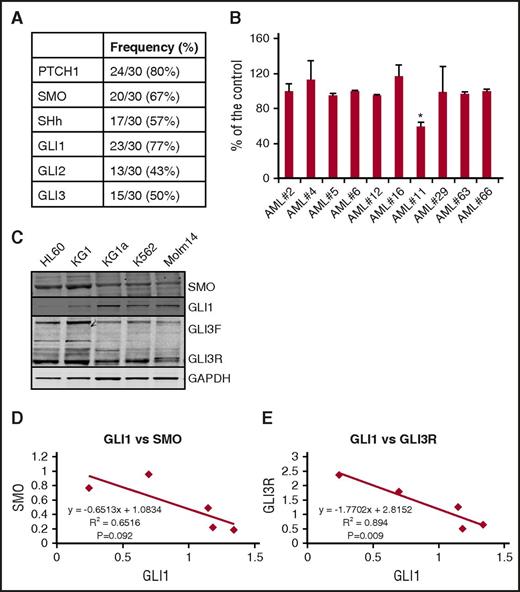
We began by profiling the gene expression of key Hh pathway members in 30 AML patient samples by using real-time Q-PCR. The majority of the AML samples expressed several Hh pathway components (SHh [17 of 30 samples, 57%], PTCH1 [24 of 30 samples, 80%], SMO [20 of 30 samples, 67%] and GLI1 [23 of 30 samples, 77%], Gli2 [13 of 30 samples, 43%], and GLI3 [15 of 30 samples, 50%]) (Figure 1A). The expression of PTCH1 and GLI1, which are transcriptional targets of the pathway, is clear evidence that the pathway is active in AML and confirms what others have reported.21 We next treated a subset of these primary patient samples, selected for expression of the target gene SMO and evidence of active Hh signaling, with PF-04449913. Consistent with prior reports, primary AML was largely resistant to SMOis, with only 1 out of 10 tested samples showing a significant response to PF-04449913 treatment (Figure 1B).
Hh signaling pathway is active in AML. (A) Expression of the Hh pathway components PTCH1, SMO, GLI1, GLI2, and GLI3 was measured by real-time Q-PCR in a panel of 30 clinical specimens from patients with AML. (B) Primary AML blasts were treated with 100 nM of the SMO antagonist PF-04449913 for 48 hours, and proliferation was measured by using XTT (Roche) assay. Results were shown as a percentage of the respective control of each sample. *P < .05 compared with control. (C) Basal SMO, GLI1, and GLI3 protein levels were measured by western blotting. Protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with GLI3 antibody. GLI3F represents the unprocessed full length GLI3, and GLI3R represents the processed repressor form of GLI3. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. (D) Correlation between GLI1 and SMO protein levels in myeloid leukemia cell lines. Protein levels were analyzed by densitometry. The x-axis represents Gli1, and the y-axis represents SMO. (E) Correlation between GLI1 and GLI3R protein levels in myeloid leukemia cell lines. Protein levels were analyzed by densitometry. The x-axis represents GLI1, and the y-axis represents GLI3R.
Hh signaling pathway is active in AML. (A) Expression of the Hh pathway components PTCH1, SMO, GLI1, GLI2, and GLI3 was measured by real-time Q-PCR in a panel of 30 clinical specimens from patients with AML. (B) Primary AML blasts were treated with 100 nM of the SMO antagonist PF-04449913 for 48 hours, and proliferation was measured by using XTT (Roche) assay. Results were shown as a percentage of the respective control of each sample. *P < .05 compared with control. (C) Basal SMO, GLI1, and GLI3 protein levels were measured by western blotting. Protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with GLI3 antibody. GLI3F represents the unprocessed full length GLI3, and GLI3R represents the processed repressor form of GLI3. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. (D) Correlation between GLI1 and SMO protein levels in myeloid leukemia cell lines. Protein levels were analyzed by densitometry. The x-axis represents Gli1, and the y-axis represents SMO. (E) Correlation between GLI1 and GLI3R protein levels in myeloid leukemia cell lines. Protein levels were analyzed by densitometry. The x-axis represents GLI1, and the y-axis represents GLI3R.
The 2 established modes of mutational activation of Hh signaling in cancer are either through the loss of the tumor suppressor PTCH or activating mutations of SMO.12 We examined the DNA sequences of 200 AML cases publicly available in TCGA and found no evidence of PTCH deletions or SMO mutations in AML or any other recurrent mutations in Hh pathway proteins (GLI1, GLI2, GLI3, and SUFU) that could account for the activation of Hh signaling observed in AML.24 The lack of response to SMO antagonists and the lack of mutations in PTCH or SMO suggest that in AML, activation of Hh targets does not occur through normal signaling function and instead occurs through mechanisms downstream of SMO activity.
To better characterize Hh pathway activity in AML, we examined a series of AML cell lines (HL60, KG1, KG1a, K562, and Molm14) for Hh pathway members. AML cell lines displayed a range of GLI1 protein levels (Figure 1C). Interestingly, all cells lines expressed SMO, however, levels of SMO seemed to be inversely correlated with GLI1 levels (Figure 1D). Furthermore, GLI1 levels also appeared to be inversely correlated with GLI3R levels (Figure 1E). Because GLI3R is a negative regulator of GLI1 expression and Hh pathway activity, these findings suggest that GLI3R could be the primary regulator of Hh target genes in AML.
Response to the SMO antagonist PF-04449913 varies among AML cell lines and is associated with expression of GLI3
To understand why PF-04449913 seemed to have limited activity against primary AML, we sought to identify SMOi-sensitive and -resistant cells lines for additional study. We treated 8 cells lines with 100 nM of PF-04449913 for 48 hours and measured changes in cell growth. Interestingly, 6 out of 8 cell lines, including Molm-14, K562, and KG1a, the cells with the highest GLI activity showed no growth inhibition with PF-04449913, whereas KG1 and HEL cells showed an ∼50% and 20% decrease in cell numbers, respectively (Figure 2A; supplemental Figure 1A). In addition, we observed that KG1 cells are similarly sensitive to SHh (supplemental Figure 2). These responses corresponded to what was seen at the level of GLI1 transcripts, with no change in the unresponsive cell lines, but a decrease in KG1 and HEL cells (Figure 2B; supplemental Figure 1B). We chose KG1 (low GLI1/high GLI3 protein levels), K562, and KG1a (high GLI1/low GLI3 protein levels) cell lines for additional study. We first confirmed that the cells had active Hh signaling by utilizing a GLI-responsive luciferase reporter construct. All the cell lines demonstrated GLI reporter activity above the baseline of the PGL3B control plasmid, with levels that corresponded to the levels of GLI1 protein and gene expression (Figure 2C). Keeping in mind that, after binding to a receptor, many receptor antagonists lead to receptor deactivation and degradation, we examined the protein levels of SMO after PF-04449913 treatment. Western analysis revealed no change in SMO levels after treatment with PF-04449913 (Figure 2D). These data demonstrate that some AML cells are inherently resistant to SMO antagonists despite evidence of SMO expression and GLI1 activity and suggest that GLI1 is regulated independently of SMO.
Response to SMO antagonist PF-04449913 varies among AML cell lines. (A) AML cell lines were treated with 100 nM of PF-04449913 for 48 hours. Cell viability was tested by trypan blue exclusion assay. *P < .05 vs vehicle-treated cells. Treatment with PF-04449913 decreased cell viability in KG1 cells with no change in K562 and KG1a cells. (B) GLI1 transcript levels, which are an indicator of Hh pathway activity, were measured by Q-PCR. GLI1 transcript levels were reduced in KG1 cells only with PF-04449913 treatment. (C) AML cell lines (KG1, KG1a, and K562) were transiently transfected with a GLI-responsive luciferase reporter or pGL3-basic, a plasmid lacking GLI binding site, as a negative control. Relative luciferase levels were measured at 48 hours posttransfection. (D) SMO protein levels were measured in cell lines treated with PF-04449913. (E) K562 and KG1 cells were transiently transfected with SMO siRNA, respectively. Non-specific scrambled siRNA (NS siRNA) was used as control. Cell viability was tested by trypan blue exclusion assay. Transfection efficiency of SMO siRNA transfection efficiency was measured by western blotting for SMO protein levels. GLI1 and GLI3 protein levels were also measured by western blotting. (F) K562 cells were transiently transfected with GLI1 siRNA. NS scrambled siRNA was used as control. Cell viability was tested by trypan blue exclusion assay. *P < .05 vs NS siRNA control. Transfection efficiency of GLI1 siRNA was measured by western blotting using GLI1 antibody. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control in panels D-F.
Response to SMO antagonist PF-04449913 varies among AML cell lines. (A) AML cell lines were treated with 100 nM of PF-04449913 for 48 hours. Cell viability was tested by trypan blue exclusion assay. *P < .05 vs vehicle-treated cells. Treatment with PF-04449913 decreased cell viability in KG1 cells with no change in K562 and KG1a cells. (B) GLI1 transcript levels, which are an indicator of Hh pathway activity, were measured by Q-PCR. GLI1 transcript levels were reduced in KG1 cells only with PF-04449913 treatment. (C) AML cell lines (KG1, KG1a, and K562) were transiently transfected with a GLI-responsive luciferase reporter or pGL3-basic, a plasmid lacking GLI binding site, as a negative control. Relative luciferase levels were measured at 48 hours posttransfection. (D) SMO protein levels were measured in cell lines treated with PF-04449913. (E) K562 and KG1 cells were transiently transfected with SMO siRNA, respectively. Non-specific scrambled siRNA (NS siRNA) was used as control. Cell viability was tested by trypan blue exclusion assay. Transfection efficiency of SMO siRNA transfection efficiency was measured by western blotting for SMO protein levels. GLI1 and GLI3 protein levels were also measured by western blotting. (F) K562 cells were transiently transfected with GLI1 siRNA. NS scrambled siRNA was used as control. Cell viability was tested by trypan blue exclusion assay. *P < .05 vs NS siRNA control. Transfection efficiency of GLI1 siRNA was measured by western blotting using GLI1 antibody. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control in panels D-F.
To investigate the possibility of SMO-independent GLI activity in AML, we performed siRNA mediated knockdown of SMO and assessed its effects on cell growth and GLI protein levels. Knockdown of SMO in the PF-04449913–resistant K562 cells did not inhibit cell proliferation or change levels of GLI1 or GLI3 (Figure 2E). In contrast, knockdown of SMO in the PF-04449913–responsive cell line KG1 led to a decrease in KG1 cell proliferation and a concomitant decrease in GLI1 protein levels. SMO knockdown did not change the levels of GLI3R, suggesting more complex regulation of GLI3 involving proteins, such as SUFU, protein kinase A, and GSK3. However, direct targeting of GLI1 by siRNA in K562 cells did lead to decreased growth, demonstrating that although GLI1 is independent of SMO and not modulated by PF-04449913 in some AML cells, GLI1 is still an important therapeutic target (Figure 2F).
GLI3R is the critical regulator of GLI1 and Hh target genes in AML
Inhibition of SMO with cyclopamine is known to inhibit pathway activity in part by increasing GLI3R, thus causing repression of Hh target genes, and in some tissues, GLI3R is absolutely required for cyclopamine to function.29,30 To date, most studies of Hh signaling in AML have focused on the upstream activator SMO or the target genes GLI1 or GLI2, but little is known about GLI3 in AML. Therefore, to better understand how Hh targets were being activated in AML and to explore the role of GLI3 in SMOi resistance, we examined levels of GLI3 in PF-04449913–responsive and –resistant AML samples.
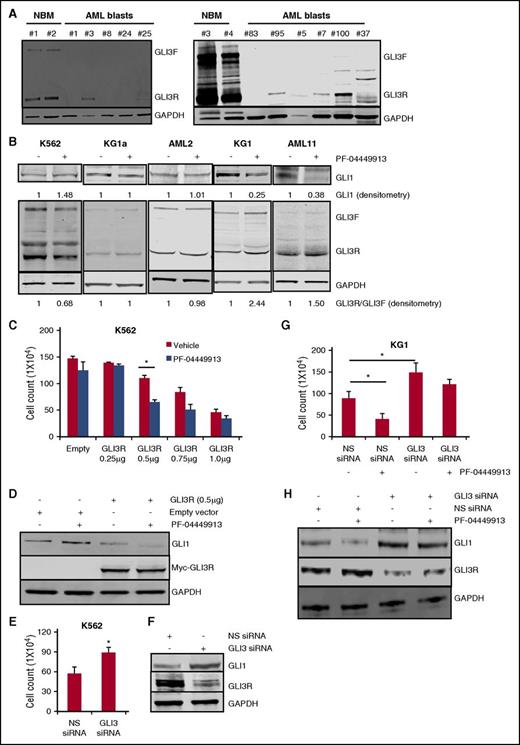
As seen in Figure 1C, cell lines with high levels of GLI1 all had low levels of GLI3. We next profiled GLI3 levels in normal bone marrow and primary AML samples. Robust expression of both GLI3F and GLI3R were detected in normal bone marrow, high levels were detected in only 1 primary AML sample, with very low levels detected in another 4 out of 10 samples. Half of the samples had absolutely no detectable GLI3 protein. In contrast, GLI1 expression was noted in all 5 patient samples tested at levels higher than that seen in normal bone marrow (Figure 3A; supplemental Figure 3). These data suggest that loss of GLI3 expression is common in AML. We next examined levels of GLI1, GLI3F, and GLI3R in a group of AML cells that showed PF-04449913 sensitivity (KG1, HEL, and AML11) and in cells resistant to PF-04449913 (K562, KG1a, ML-2, MV4;11, Molm14, U937, and AML2). K562 cells demonstrated a paradoxical decrease in GLI3R levels with PF-04449913 treatment that was associated with an increase in GLI1 levels (Figure 3B; supplemental Figure 4). Most of the AML cell lines and AML2 primary patient samples showed no changes in GLI3 or GLI1 on treatment with PF-04449913. In contrast, treatment with PF-04449913 in the SMOi-sensitive group (KG1, HEL, and AML11) demonstrated the expected increases in GLI3R with a reciprocal decrease in GLI1 (Figure 3B; supplemental Figure 4). These data confirm that sensitivity to SMOis is associated with changes in GLI3.
GLI3 is downregulated in AML and GLI3R is a critical regulator of Hh signaling in AML. (A) Western blotting was used to measure GLI3 protein levels in clinical specimens from patients with AML and normal bone marrow (NBM). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Compared with NBM, AML samples showed significantly lower GLI3 levels. (B) AML cell lines and primary AML blasts were treated with PF-04449913, and cell lysates were analyzed for GLI1 and GLI3 protein levels. SMO antagonist increased the GLI3R:GLI3F ratio in responsive KG1 cells and primary AML11 blasts with no change in nonresponsive K562 and KG1a cells and primary AML2 blasts. PF-04449913 decreased GLI1 levels in KG1 and AML11 cells as shown by GLI1 densitometry. GAPDH was used as a loading control in all experiments. Densitometry analysis was done by using Image Studio Lite software. (C) K562 cells were transiently transfected with empty and Myc-tagged GLI3R constructs (0, 0.25, 0.5, 0.75, and 1 μg) followed by PF-04449913 treatment. Forced expression of GLI3R at 0.5 μg significantly reduced cell viability, which was additionally reduced after PF-04449913 treatment. *P < .05. (D) After cells were transfected with the indicated constructs and treated with PF-04449913, cell lysates were prepared and subjected to immunoblotting with anti-GLI1, anti-Myc, and anti-GAPDH antibodies. (E-F) K562 cells were transiently transfected with iLenti-si-Gli3 siRNA. NS scrambled siRNA was used as control. Cell viability was tested by trypan blue exclusion assay. *P < .05 vs NS siRNA control. Transfection efficiency of GLI3 siRNA was measured by western blotting by using GLI3 antibody. (G- H) KG1 cells were transfected with iLenti-si-Gli3 siRNA followed by treatment with PF-04449913. NS scrambled siRNA was used as control. Cell viability was tested by trypan blue exclusion assay. *P < .05. GLI1 and GLI3 protein levels were measured by western blotting. GAPDH was used as a loading control.
GLI3 is downregulated in AML and GLI3R is a critical regulator of Hh signaling in AML. (A) Western blotting was used to measure GLI3 protein levels in clinical specimens from patients with AML and normal bone marrow (NBM). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Compared with NBM, AML samples showed significantly lower GLI3 levels. (B) AML cell lines and primary AML blasts were treated with PF-04449913, and cell lysates were analyzed for GLI1 and GLI3 protein levels. SMO antagonist increased the GLI3R:GLI3F ratio in responsive KG1 cells and primary AML11 blasts with no change in nonresponsive K562 and KG1a cells and primary AML2 blasts. PF-04449913 decreased GLI1 levels in KG1 and AML11 cells as shown by GLI1 densitometry. GAPDH was used as a loading control in all experiments. Densitometry analysis was done by using Image Studio Lite software. (C) K562 cells were transiently transfected with empty and Myc-tagged GLI3R constructs (0, 0.25, 0.5, 0.75, and 1 μg) followed by PF-04449913 treatment. Forced expression of GLI3R at 0.5 μg significantly reduced cell viability, which was additionally reduced after PF-04449913 treatment. *P < .05. (D) After cells were transfected with the indicated constructs and treated with PF-04449913, cell lysates were prepared and subjected to immunoblotting with anti-GLI1, anti-Myc, and anti-GAPDH antibodies. (E-F) K562 cells were transiently transfected with iLenti-si-Gli3 siRNA. NS scrambled siRNA was used as control. Cell viability was tested by trypan blue exclusion assay. *P < .05 vs NS siRNA control. Transfection efficiency of GLI3 siRNA was measured by western blotting by using GLI3 antibody. (G- H) KG1 cells were transfected with iLenti-si-Gli3 siRNA followed by treatment with PF-04449913. NS scrambled siRNA was used as control. Cell viability was tested by trypan blue exclusion assay. *P < .05. GLI1 and GLI3 protein levels were measured by western blotting. GAPDH was used as a loading control.
To prove that changes in GLI3R were sufficient to affect AML growth and GLI1 levels, we expressed GLI3R in SMOi-resistant K562 and KG1a cells (which have low levels of GLI3R) and performed knockdown experiments of GLI3 in K562 cells. As expected, expression of GLI3R in K562 and KG1a led to decreased cell growth and decreased GLI1 levels (Figure 3C-D; supplemental Figure 6). Furthermore, GLI3R expression reversed the paradoxical increase in GLI1 observed in K562 cells treated with PF-04449913, suggesting that GLI3R expression altered responsiveness to SMOis (Figure 3D). In KG1a cells, transduction with GLI3R plasmid caused modest suppression of growth compared with K562 cells, but demonstrated significant growth inhibition on PF-04449913 treatment at 0.5 μg and 1 μg of GLI3R plasmid (supplemental Figure 6). In contrast, siRNA-mediated knockdown of GLI3 in K562 cells led to an increase in cell growth and GLI1, consistent with the role of GLI3 as a negative regulator of growth and GLI1 levels in SMOi-resistant AML (Figure 3E-F).
We next examined the effects of GLI3 loss on the SMOi-sensitive KG1 cell line. As in K562, loss of GLI3 leads to an increase in GLI1 and cell growth (Figure 3G). However, loss of GLI3 rendered the KG1 cells relatively insensitive to the cytotoxic effect of PF-04449913, despite the fact that GLI1 levels fell with PF-04449913 treatment (Figure 3H). These data suggest that in SMOi-sensitive cells, SMO activity may continue to control GLI1 levels in the absence of GLI3, however, it is the direct inhibition of GLI3R targets, other than GLI1, that mediate the tumor suppressor effects of GLI3R.
AKT is a target of GLI3R tumor suppressor activity
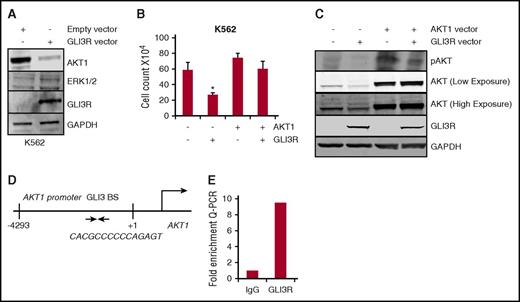
Previous studies have reported crosstalk between Hh signaling and survival pathways involving extracellular signal-regulated kinase (ERK) and AKT, and therefore, we hypothesized that GLI3 might interact with these pathways in leukemia.11,31,32 To explore this possibility, we expressed GLI3R in leukemia cells and measured changes in ERK and AKT levels. Expression of GLI3R significantly decreased total AKT levels, with very little change in ERK levels (Figure 4A). Given the role of AKT in several oncogenic pathways, we hypothesized that AKT might be a critical target of GLI3R tumor suppressor activity. Indeed, enforced expression of a constitutively active form of AKT rescued the cell proliferation defect caused by GLI3R overexpression (Figure 4B-C). These results implicate AKT as the critical mediator of GLI3R tumor suppressor function.
GLI3R suppresses Hh signaling via transcriptional repression of AKT. (A) K562 cells were transiently transfected with the GLI3R expression construct. After 48 hours, cell lysates were separated and immunoblotted using anti-AKT, anti-ERK1/2, anti-Myc, and anti-glyceraldehyde-3-phosphate dehydrogenase antibodies. Total AKT levels were significantly downregulated in GLI3R-expressing K562 cells. (B-C) Expression of AKT in GLI3R-overexpressing cells rescues the GLI3R-mediated growth inhibition. K562 cells were transiently transfected with the GLI3R and/or the AKT1 expression construct. Cell viability was tested by using trypan blue exclusion assay. *P < .05 compared with control. (D) Schematic diagram of the AKT1 promoter region with potential GLI3 binding site (BS). The 9 bp GLI-binding site sequence is shown. (E) Fold enrichment of GLI3 on AKT. ChIP assay was performed with control immunoglobulin G and GLI3R-specific antibodies.
GLI3R suppresses Hh signaling via transcriptional repression of AKT. (A) K562 cells were transiently transfected with the GLI3R expression construct. After 48 hours, cell lysates were separated and immunoblotted using anti-AKT, anti-ERK1/2, anti-Myc, and anti-glyceraldehyde-3-phosphate dehydrogenase antibodies. Total AKT levels were significantly downregulated in GLI3R-expressing K562 cells. (B-C) Expression of AKT in GLI3R-overexpressing cells rescues the GLI3R-mediated growth inhibition. K562 cells were transiently transfected with the GLI3R and/or the AKT1 expression construct. Cell viability was tested by using trypan blue exclusion assay. *P < .05 compared with control. (D) Schematic diagram of the AKT1 promoter region with potential GLI3 binding site (BS). The 9 bp GLI-binding site sequence is shown. (E) Fold enrichment of GLI3 on AKT. ChIP assay was performed with control immunoglobulin G and GLI3R-specific antibodies.
AKT is an established transcriptional target of GLI1, but there are no reports on the role of GLI3 in regulating AKT expression.33 GLI3R could directly control AKT expression through binding to GLI binding sites in the AKT promoter, or, because GLI3R regulates GLI1 levels, it is possible that GLI3R was indirectly regulating AKT via GLI1. To confirm that GLI3R targets AKT by directly binding to the promoter region, we performed a ChIP–Q-PCR assay for GLI3R. K562 cells were transfected with the GLI3R-Myc construct, and DNA was immunoprecipitated with an anti-Myc antibody. Q-PCR using primers for binding sites analyzed genomic DNA fragments bound to GLI3R; binding site (–4293/+1) that include conserved GLI binding elements in the AKT promoter region (Figure 4D). Ten-fold enrichment for the AKT promoter was seen with immunoprecipitation of GLI3R compared with immunoglobulin G control (Figure 4E). These results demonstrate that GLI3R directly interacts with the AKT promoter to repress gene expression.
Decitabine reactivates GLI3 and alters Hh pathway activity
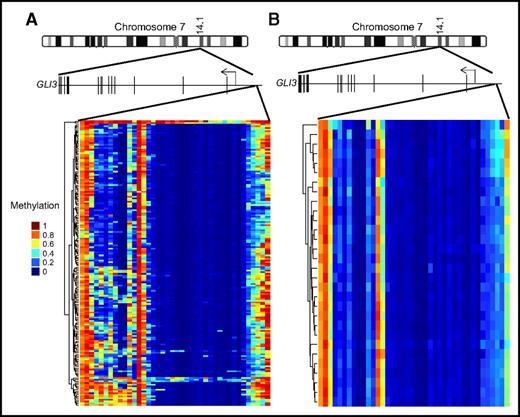
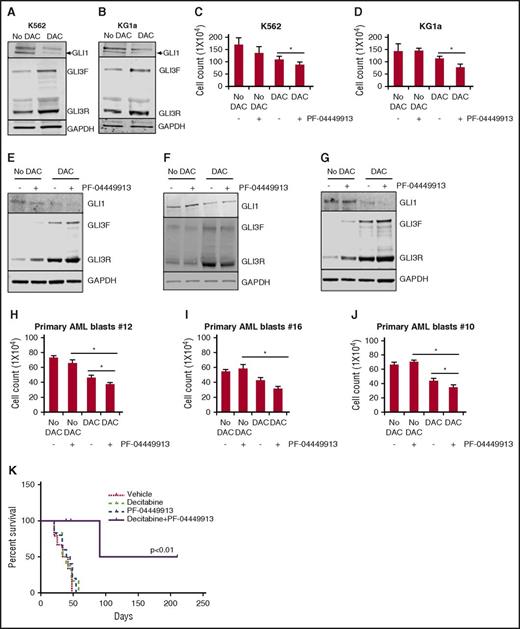
We have demonstrated that loss of GLI3 is associated with increased Hh pathway activity and resistance to SMOis. To better understand how common loss of GLI3 expression is in a larger data set, we examined gene expression profiles of AML samples from the TCGA data set and found that GLI3 expression was detectable in only 7 of 169 samples (4%).24 Focal recurrent deletions around the GLI3 locus were not detected in the same data set.24 Therefore, we speculated that GLI3 might be silenced epigenetically in AML. We examined the GLI3 promoter regions of all known transcriptional isoforms for evidence of abnormal methylation in the same TCGA data set. Using normal bone marrow stem cells and progenitor fractions as comparators, we observed areas of hypermethylation in the GLI3 promoter of AML, which was not seen in controls24,25 (Figure 5). Therefore, we hypothesized that hypermethylation could be a potential mechanism for GLI3 inactivation, and re-expression of silenced GLI3 using demethylating agents could alter Hh signaling activity. To test this hypothesis, we treated K562 and KG1a cells with DAC and observed an increase in GLI3 with a corresponding decrease in GLI1 and reductions in cell growth (Figure 6A-D). We then treated 3 primary AML samples that were resistant to PF-04449913 and observed a similar pattern of increased GLI3R and modulation of GLI1 (Figure 6E-G). Combined treatment with DAC and PF-04449913 led to additional increases in GLI3R and modulation of GLI1 that corresponded to additional decreases in leukemia cell survival (Figure 6H-J). Finally, we treated immunodeficient mice xenografted with K562 cells with DAC, PF-04449913, or a combination of both and monitored mice for survival. Treatment with DAC and PF-04449913 in combination was well tolerated and significantly prolonged the survival of mice, suggesting synergy of the combination (Figure 6K).
GLI3 is hypermethylated in AML. Schematic representation of the GLI3 gene (15 exons; not drawn to scale) on chromosome 7. (A) GLI3 promoter methylation data from the TCGA AML cohort. (B) GLI3 promoter methylation data from a cohort of normal bone marrow stem and progenitor cells.
GLI3 is hypermethylated in AML. Schematic representation of the GLI3 gene (15 exons; not drawn to scale) on chromosome 7. (A) GLI3 promoter methylation data from the TCGA AML cohort. (B) GLI3 promoter methylation data from a cohort of normal bone marrow stem and progenitor cells.
Epigenetic reactivation of GLI3 increases sensitivity to the SMO antagonist PF-04449913 in AML. (A-B) Treatment with the demethylating agent DAC increases GLI3R protein levels in AML cell lines. K562 and KG1a cells (low basal GLI3 levels) were cultured in the presence of DAC (1 nM) for 5 days with the drug replaced daily, and cell lysates were subjected to immunoblotting for GLI3 levels. Glyceraldehyde-3-phosphate dehydrogenase was used as loading control. (C-D) K562 and KG1a cells were treated with DAC (1 nM) for 5 days followed by PF-04449913 (100 nM) treatment for 2 days. Cell proliferation was measured by using trypan blue assay. *P < .05. (E-J) Primary AML blasts were treated ex vivo with DAC and PF-04449913. DAC sensitizes AML blasts to PF-04449913 by reducing cell viability and downregulating GLI1 expression. *P < .05. (K) Kaplan-Meier survival curve of mice transplanted with K562 cells and treated with vehicle (n = 6), PF-04449913 (n = 5), DAC alone (n = 6), and a combination of PF-04449913 and DAC (n = 6).
Epigenetic reactivation of GLI3 increases sensitivity to the SMO antagonist PF-04449913 in AML. (A-B) Treatment with the demethylating agent DAC increases GLI3R protein levels in AML cell lines. K562 and KG1a cells (low basal GLI3 levels) were cultured in the presence of DAC (1 nM) for 5 days with the drug replaced daily, and cell lysates were subjected to immunoblotting for GLI3 levels. Glyceraldehyde-3-phosphate dehydrogenase was used as loading control. (C-D) K562 and KG1a cells were treated with DAC (1 nM) for 5 days followed by PF-04449913 (100 nM) treatment for 2 days. Cell proliferation was measured by using trypan blue assay. *P < .05. (E-J) Primary AML blasts were treated ex vivo with DAC and PF-04449913. DAC sensitizes AML blasts to PF-04449913 by reducing cell viability and downregulating GLI1 expression. *P < .05. (K) Kaplan-Meier survival curve of mice transplanted with K562 cells and treated with vehicle (n = 6), PF-04449913 (n = 5), DAC alone (n = 6), and a combination of PF-04449913 and DAC (n = 6).
Discussion
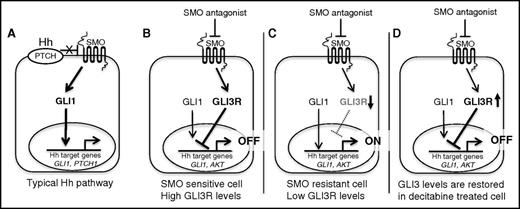
Aberrant Hh activation has been implicated in tumorigenesis for a wide variety of tumors.34 Although SMO antagonists have been very successful in advanced BCC, these inhibitors have failed to show any clinical activity in other solid tumors, including pancreatic, ovarian,35 and colorectal cancer.36 For most cancers, including BCC, GLI1 expression has been the primary criterion for selecting cancers with Hh activity, and its modulation has been the primary biomarker of treatment response.37-39 This approach is based on the typical model of Hh signaling, which puts GLI1 at the center of the signaling response (Figure 7A). In this article, we revise that model to emphasize the critical role of GLI3R in regulating the Hh response in AML. Although the role of GLI3 as a negative regulator of Hh signaling is well established in the context of normal development, its role in cancer has largely been ignored. Our data confirm previous reports showing that the vast majority of AMLs have evidence of Hh target gene activation. We also demonstrate that GLI3 expression is epigenetically silenced in most AMLs. We believe that most of the Hh pathway activity observed in AML is primarily a result of loss of GLI3R suppression rather than SMO-based activation. This is supported by our observation that the majority of AMLs are resistant to SMOis, although some do demonstrate sensitivity. In AML cells with active Hh signaling and adequate levels of GLI3R, we believe that SMO antagonists exert most of their antitumor activity through increases in GLI3R levels (Figure 7B). In most AMLs, however, GLI3 is epigenetically silenced, and therefore SMO antagonists are ineffective (Figure 7C). Treatment of AML with hypomethylating agents, such as DAC, restores GLI3 expression, which leads to suppression of Hh pathway activity (Figure 7D). Our data suggest GLI3R as a candidate biomarker for selecting patients who will be responsive to SMOis and for pharmacodynamic monitoring of patients being treated with SMOis. We also provide a clear mechanistic rationale for combining hypomethylator agents and SMO antagonists in AML.
Schematic of canonical Hh pathway. (A) SMO antagonist–responsive cell (eg, BCC). (B-D) Revised schematic of Hh signaling in SMO-sensitive, SMO-resistant, and decitabine-treated AML cells.
Schematic of canonical Hh pathway. (A) SMO antagonist–responsive cell (eg, BCC). (B-D) Revised schematic of Hh signaling in SMO-sensitive, SMO-resistant, and decitabine-treated AML cells.
An important limitation of our work is that we do not address the potential role of tumor/stromal interactions on Hh signaling and the possible therapeutic benefit of inhibiting Hh signaling in tumor stroma. Our data suggest that the majority of AML cells are inherently resistant to SMOis, although recent clinical trials demonstrate a survival benefit in patients treated with PF-04449913. Inhibition of stromal Hh signaling may explain the unexpected degree of efficacy seen in clinical trials.
Further complicating our understanding of Hh signaling in leukemia is our recent observation that primary cilia are lost on most leukemia cells, despite being present on all normal blood and bone marrow cells.40 Our report shows that KG1 cells, which are sensitive to SMOis, have the highest number of primary cilia, whereas the SMOi-insensitive cell lines (KG1a and K562) show very few primary cilia.40 These findings may explain the different effects of SMO knockdown vs SMO inhibition on GLI3R levels. The knockdown depletes SMO from the cells, whereas PF-04449913 has no effects on SMO levels, but instead prevents SMO localization to cilia. The paradoxical changes in GLI3R seen in K562 cells may be due to aberrant noncilial activity of SMO in leukemia cells. A comprehensive examination of primary cilia from patients treated with PF-04449913 is needed to validate the presence of primary cilia as a biomarker for SMO sensitivity.
In the present study, we also provide the first evidence, to our knowledge, that GLI3R is a potential tumor suppressor and results in the repression of AKT expression by directly binding to its promoter region. However, AKT itself has been shown to stimulate the transcriptional activity and nuclear localization of GLI1. In addition, AKT has been shown to enhance GLI protein stability and SMO-independent activation of GLI through transforming growth factor-β, endothelial growth factor receptor, and RAS/RAF/MEK/ERK pathways has been reported.41-43 Others have shown that phosphatidylinositol 3-kinase/AKT contributes to activation of the Hh/GLI1 signaling pathway via GSK3β and ERK1/2.44,45 Therefore, any analysis of the respective roles of GLI3R and AKT regulation of Hh targets is confounded by negative and positive feedback loops with GLI1, which is a limitation of this study.
Beyond AKT, the Hh pathway has hundreds of putative target genes that could be regulated by GLI3 and have important functions in leukemia. We have undertaken a comprehensive analysis of GLI3 target genes within hematopoiesis and leukemia to better understand GLI3 function in leukemia that we hope will further elucidate the function of GLI3 in cancer. Important questions regarding the transcriptional regulation of GLI3 and its proteolytic processing to repressor form in the context of leukemia remain unanswered. Ultimately, effective targeting of the Hh pathway in leukemia and other cancers may require direct targeting of the GLI proteins and GLI3 in particular.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alan Epstein (University of Southern California) for the HEL and ML-2 cell lines.
This work was supported in part by award P30CA014089 and by career development award K08 CA154975-01A1 (A.A.M.) from the National Institutes of Health, National Cancer Institute.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Authorship
Contribution: P.C. designed and performed the experiments, analyzed data, and prepared the manuscript; M.S. performed the experiments; T.J.T. and M.G. analyzed data; and A.A.M. designed the research, analyzed data, and prepared the manuscript.
Conflict-of-interest disclosure: A.A.M. received research funding and served as a consultant for Pfizer, Inc. The remaining authors declare no competing financial interests.
Correspondence: Akil A. Merchant, Division of Hematology, Department of Medicine, USC Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, 1450 Biggy St, Norris Research Tower 3501, Los Angeles, CA 90033; e-mail: akil.merchant@med.usc.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal